
Neural networks made easy (Part 9): Documenting the work
Contents
- Introduction
- 1. The basic principles of creating documentation
- 2. Selecting tools
- 3. Documenting in the code
- 4. Preparation in the code source file
- 5. Generating documentation
- Conclusions
- References
- Programs Used in the Article
Introduction
During previous eight articles, we have been adding new objects and expanding the functionality of existing ones. , we have been adding new objects and expanding the functionality of existing ones. All these additions expanded our library. We have also added an OpenCL program file. Now the code is 10 times larger than the first one. It is becoming difficult to trace the relationships between objects in the code. The readers may find the code very confusing and difficult to understand. I try to provide a detailed description of action logics in each article. But the demonstration of separate action chains does not provide a general understanding of the program.
That is why I decided to demonstrate the creation of documentation to code, which would allow to look at the code from another perspective. The purpose of the documentation is to generalize all objects and methods in the library and to build a hierarchy of inheritance of objects and methods. This should give us a general idea of what we have done.
1. The basic principles of creating documentation
What is the purpose of technical documentation in IT developments? First of all, the documentation gives a general idea of the program architecture and operation. Proper documentation allows development teams to correctly distinguish areas of responsibility, to track all changes in the code and to evaluate their influence on the entire algorithm and architecture integrity. It also facilitates knowledge sharing. Understanding the integrity of the program architecture makes it possible to analyze and to work out ways of project development.
Properly written technical documentation should take into account the qualifications of its target user. The information should be clear and should avoid excessive explanations. Documentation should contain all the information the user needs. At the same time, it should be concise and easy to read. Excessive content takes extra time to read and annoys the reader. It is even more annoying if the user reads lengthy documentation and cannot find the required information. This leads to the next rule: documentation must have convenient tools for information search. A user-friendly interface and cross-references make it easy to find the information you need.
The documentation should contain the complete architecture of the solution and a description of the implemented technical solutions. The complete and detailed solution description facilitates the development and further support. And it is very important to always keep the documentation up to date. Outdated information can lead to contradictory management decisions and, as a result, it can unbalance the entire development.
Also, the documentation must necessarily describe the interfaces between the components.
2. Selecting tools
There are some specialized programs which can assist in creating documentation. I think, the most common ones are Doxygen, Sphinx, Latex (there are also some other tools). All of them aim at reducing labor costs for creating documentation. Of course, each program was created by developers to solve specific problems. For example, Doxygen is a program for creating documentation for C++ programs and similar programming languages. Sphinx was created for documentation for Python. But this does not mean that they are highly specialized in programming languages. Both of these programs work well with various programming languages. The relevant program websites provide detailed reference on how to use them, so you can choose the one that suits you best.
Documentation for MQL5 was already discussed earlier, in the article "Automatic creation of documentation for MQL5 programs". This article suggested using Doxygen. I also use this program for my developments. MQL5 syntax is close to C++ and so Doxygen is quite suitable for MQL5 programs. I like the fact that in order to create documentation, you only need to add appropriate comments to program code, while the specialized software will do the rest. Moreover, Doxygen allows inserting hyperlinks and mathematical formulas, which is important given the topic of the articles. We will consider the functionality usage specifics further in this article, using specific examples.
3. Documenting in the code
As mentioned above, to generate documentation, you need to add comments in the program code. Doxygen creates documentation based on these comments. Naturally, not all code comments should be included in the documentation. Some of comments may contain developer notes, somewhere commenting is added for unused code. Doxygen developers have provided ways to mark comments to be included in documentation. There are several options, and you can choose the one which is convenient for you.
Similarly to MQL5, comments for documentation can be single-line and multi-line. In order not to interfere with the direct code use in the future, we will use the standard options for inserting comments, and we will use an additional slash for single-line comments or an asterisk for multi-line comments. Optionally, an exclamation mark can be used to identify comment blocks for documentation.
/// A single-line comment for documentation /** A multi-line block for documentation */ //! An alternative single-line comment for documentation /*! An alternative multi-line block for documentation */
Please note that a multi-line comment block does not mean that the same multi-line presentation will be used in documentation. If you need to separate the brief and detailed description of a program object, you can add different comment blocks or use special commands, which are indicated by the character "\" or "@". Command "\n" can be used for forced line break.
Option 1: Separate blocks /// Short description /** Detailed description */ Option 2: Use of special commands /** \brief Brief description \details Detailed description */
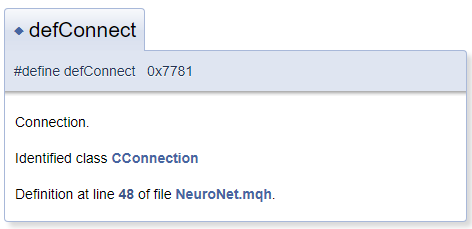
In general, it is assumed that the documentation object is located in the file next to the comment block. But in practice, it can be needed to comment on the object located before the comment block. In this case, use character "<" which informs Doxygen that the commented object is located before the block. To create cross-references in comments, precede the reference object with "#". Below is an example of code and of a block it generated in the documentation. In the generated template, "CConnection" is a reference pointing to the documentation page of the appropriate class.
#define defConnect 0x7781 ///<Connection \details Identified class #CConnection
Doxygen capabilities are extensive. The complete list of commands and their descriptions are available on the program page, under the documentation section. Furthermore, Doxygen understands HTML and XML markup. All these features allow solving a variety of tasks when creating documentation.
4. Preparation in the code source file
Now that we have reviewed the tool capabilities, we can start working on the documentation. First, let us describe our files.
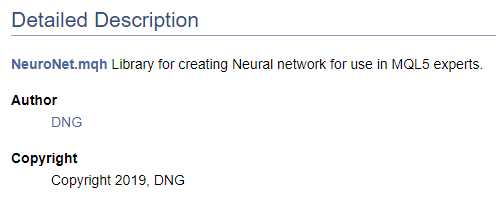
/// \file /// \brief NeuroNet.mqh /// Library for creating Neural network for use in MQL5 experts /// \author [DNG](https://www.mql5.com/en/users/dng) /// \copyright Copyright 2019, DNG
and
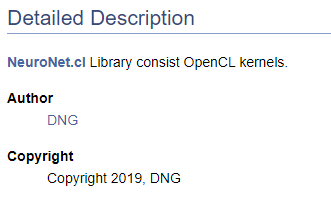
/// \file /// \brief NeuroNet.cl /// Library consist OpenCL kernels /// \author <A HREF="https://www.mql5.com/en/users/dng"> DNG </A> /// \copyright Copyright 2019, DNG
Pay attention that in the first case the \author pointer is followed by the markup provided by Doxygen, and in the second case the HTML markup is used. This is used here to demonstrate different options for creating hyperlinks. The result is the same in these cases - it creates a link to my profile at MQL5.com.
Of course, when starting the creation of code documentation, it is necessary to have at least a high-level structure of the desired result. The understanding of the final structure enables a correct grouping of documentation objects. Let us combine the created enumerations into a separate group. To declare a group, use the "\defgroup" command. The boundaries of the group are denoted by characters "@{" and "@}".
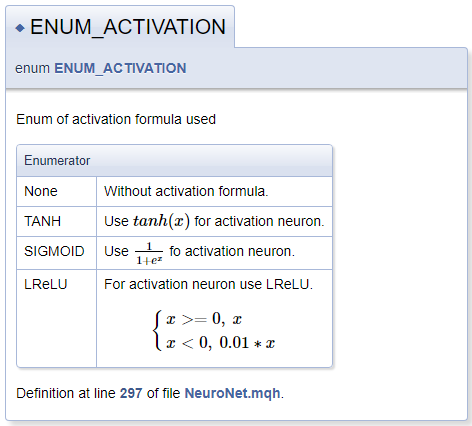
///\defgroup enums ENUM ///@{ //+------------------------------------------------------------------+ /// Enum of activation formula used //+------------------------------------------------------------------+ enum ENUM_ACTIVATION { None=-1, ///< Without activation formula TANH, ///< Use \f$tanh(x)\f$ for activation neuron SIGMOID, ///< Use \f$\frac{1}{1+e^x}\f$ fo activation neuron LReLU ///< For activation neuron use LReLU \f[\left\{ \begin{array} a x>=0, \ x \\x<0, \ 0.01*x \end{array} \right.\f] }; //+------------------------------------------------------------------+ /// Enum of optimization method used //+------------------------------------------------------------------+ enum ENUM_OPTIMIZATION { SGD, ///< Stochastic gradient descent ADAM ///< Adam }; ///@}
When describing activation functions, I have demonstrated the functionality for declaring mathematical formulas by means of MathJax. Descriptions of such formulas should be placed between a pair of "\f$" commands if you wish display the formula in a text line, or between commands "\f[" and "\f]" if you want the formula to appear on a separate line. The "\frac" command allows describing a fraction. The command is followed by the numerator and denominator of the fraction in curly braces.
When describing LReLU, we needed a unifying left curly brace. To create it, we used commands "\left\{" and "\right\.". The "\right" command is followed by "\.", because the right brace is not needed in the formula. Otherwise, the period would be replaced by a closing curly brace. An array of strings is declared inside the block using commands "\begin{array} a" and "\end{array}", the separation of array elements is performed by the "\\" command. The "\ " characters allow adding a forced space.
The generated documentation block is shown below.
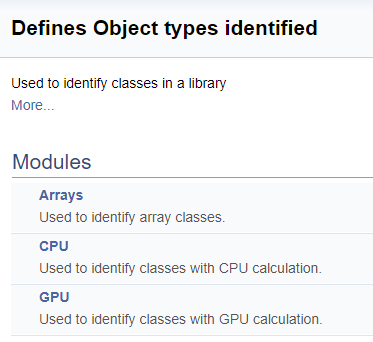
In the next step, let us create a separate group for class identifiers in the library. Inside the group, we will allocate subgroups of arrays, neurons calculating operations on CPU and neurons calculating operations on GPU. A link to the appropriate class is added as explained earlier.
///\defgroup ObjectTypes Defines Object types identified ///Used to identify classes in a library ///@{ //+------------------------------------------------------------------+ ///\defgroup arr Arrays ///Used to identify array classes ///\{ #define defArrayConnects 0x7782 ///<Array of connections \details Identified class #CArrayCon #define defLayer 0x7787 ///<Layer of neurons \details Identified class #CLayer #define defArrayLayer 0x7788 ///<Array of layers \details Identified class #CArrayLayer #define defNet 0x7790 ///<Neuron Net \details Identified class #CNet ///\} ///\defgroup cpu CPU ///Used to identify classes with CPU calculation ///\{ #define defConnect 0x7781 ///<Connection \details Identified class #CConnection #define defNeuronBase 0x7783 ///<Neuron base type \details Identified class #CNeuronBase #define defNeuron 0x7784 ///<Full connected neuron \details Identified class #CNeuron #define defNeuronConv 0x7785 ///<Convolution neuron \details Identified class #CNeuronConv #define defNeuronProof 0x7786 ///<Proof neuron \details Identified class #CNeuronProof #define defNeuronLSTM 0x7791 ///<LSTM Neuron \details Identified class #CNeuronLSTM ///\} ///\defgroup gpu GPU ///Used to identify classes with GPU calculation ///\{ #define defBufferDouble 0x7882 ///<Data Buffer OpenCL \details Identified class #CBufferDouble #define defNeuronBaseOCL 0x7883 ///<Neuron Base OpenCL \details Identified class #CNeuronBaseOCL #define defNeuronConvOCL 0x7885 ///<Convolution neuron OpenCL \details Identified class #CNeuronConvOCL #define defNeuronProofOCL 0x7886 ///<Proof neuron OpenCL \details Identified class #CNeuronProofOCL #define defNeuronAttentionOCL 0x7887 ///<Attention neuron OpenCL \details Identified class #CNeuronAttentionOCL ///\} ///@}
The division into groups in the generated documentation looks as follows.
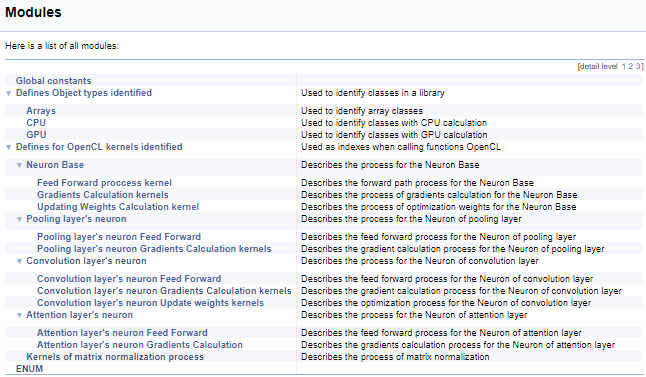
Next, we will work on a large group of definitions for working with OpenCL kernels. In this block, mnemonic names are assigned to kernel indices and their parameters, which are used when calling kernels from the main program. Using the above technology, we will split this group by the class of neurons from which the kernel is called, and then by the content of operations in the kernel (feed-forward, gradient back propagation, updating the weight coefficients). I will not provide the full code here - it is available in the attachment below. The logic for constructing subgroups is similar to the above example. The screenshot below shows the complete group structure.
Continuing with the kernels, let us move on to commenting on the OpenCL program. To create a coherent documentation structure and to get a general picture, we will use another Doxygen command "\ingroup", which allows adding new documentation objects to previously created groups. Let us use it to add kernels to the earlier created groups of indices for working with kernels. In the kernel description, add a link to the calling class and to an article on this site with a description of the process. Next, let us describe kernel parameters. The usage of pointers "[in]" and "[out]" will show the direction of the information flow. Cross-references will show the format of the data.
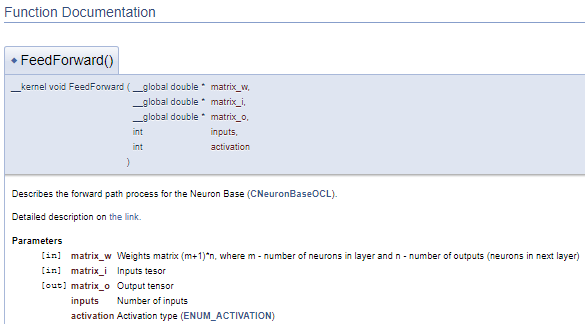
///\ingroup neuron_base_ff Feed forward process kernel /// Describes the forward path process for the Neuron Base (#CNeuronBaseOCL). ///\details Detailed description on <A HREF="https://www.mql5.com/en/articles/8435#para41">the link.</A> //+------------------------------------------------------------------+ __kernel void FeedForward(__global double *matrix_w,///<[in] Weights matrix (m+1)*n, where m - number of neurons in layer and n - number of outputs (neurons in next layer) __global double *matrix_i,///<[in] Inputs tesor __global double *matrix_o,///<[out] Output tensor int inputs,///< Number of inputs int activation///< Activation type (#ENUM_ACTIVATION) )
The above code will generate the following documentation block.
In the above example, the description of the parameters is given immediately after their declaration. But this approach can make the code clumsy. In such cases, it is suggested to use the "\param" command to describe the parameters. By using this command, we can describe parameters in any part of the file, but we need to directly specify the parameter name.
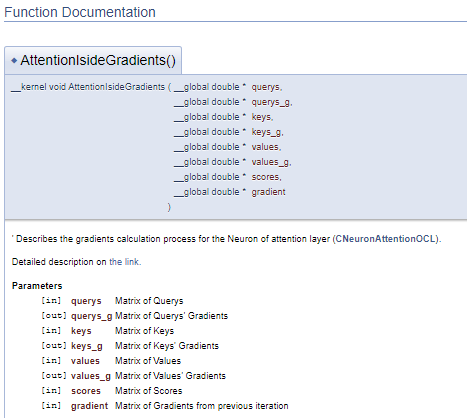
///\ingroup neuron_atten_gr Attention layer's neuron Gradients Calculation kernel /// Describes the gradients calculation process for the Neuron of attention layer (#CNeuronAttentionOCL). ///\details Detailed description on <A HREF="https://www.mql5.com/ru/articles/8765#para44">the link.</A> /// @param[in] querys Matrix of Querys /// @param[out] querys_g Matrix of Querys' Gradients /// @param[in] keys Matrix of Keys /// @param[out] keys_g Matrix of Keys' Gradients /// @param[in] values Matrix of Values /// @param[out] values_g Matrix of Values' Gradients /// @param[in] scores Matrix of Scores /// @param[in] gradient Matrix of Gradients from previous iteration //+------------------------------------------------------------------+ __kernel void AttentionIsideGradients(__global double *querys,__global double *querys_g, __global double *keys,__global double *keys_g, __global double *values,__global double *values_g, __global double *scores, __global double *gradient)
This approach generates a similar block of documentation, but it allows separating the block of comments from the program code. Thus, the code becomes easier to read.
The main work concerns documentation for our library classes and their methods. We need to describe all the classes used and their methods. To do this, we will use all the above-described commands in different variations and will add some new ones. First, let us add the class to the appropriate group, as we did it earlier with kernels (the \ingroup command). The "\class" command informs Doxygen that the below description applies to the class. In command parameters, specify the class name in order to link description to the right object
Using the "\brief" and "\details" commands, provide a brief and an extended class description. In the detailed description, add a hyperlink to the corresponding article. Here, we will add an anchor link to a specific section of the article, which will enable users to find the required information faster.
Add their descriptions directly to the variable declaration line. If necessary, add links to explanatory objects. There is no need to set pointers to the classes of declared objects in the comments, while Doxygen will add them automatically.
Similarly, describe the methods of the classes. However, unlike variables, a description of the parameters should be added in comments. To do this, use the earlier described "\param" commands along with the "[in]", "[out]", "[in,out]" pointers. Describe the method execution result using the "\return" command.
It is also possible to attach individual methods to groups by certain features. For example, they can be combined by functionality.
The below code shows all the above steps.
///\ingroup neuron_base ///\class CNeuronBaseOCL ///\brief The base class of neuron for GPU calculation. ///\details Detailed description on <A HREF="https://www.mql5.com/ru/articles/8435#para45">the link.</A> //+------------------------------------------------------------------+ class CNeuronBaseOCL : public CObject { protected: COpenCLMy *OpenCL; ///< Object for working with OpenCL CBufferDouble *Output; ///< Buffer of Output tenzor CBufferDouble *PrevOutput; ///< Buffer of previous iteration Output tenzor CBufferDouble *Weights; ///< Buffer of weights matrix CBufferDouble *DeltaWeights; ///< Buffer of last delta weights matrix (#SGD) CBufferDouble *Gradient; ///< Buffer of gradient tenzor CBufferDouble *FirstMomentum; ///< Buffer of first momentum matrix (#ADAM) CBufferDouble *SecondMomentum; ///< Buffer of second momentum matrix (#ADAM) //--- const double alpha; ///< Multiplier to momentum in #SGD optimization int t; ///< Count of iterations //--- int m_myIndex; ///< Index of neuron in layer ENUM_ACTIVATION activation; ///< Activation type (#ENUM_ACTIVATION) ENUM_OPTIMIZATION optimization; ///< Optimization method (#ENUM_OPTIMIZATION) //--- ///\ingroup neuron_base_ff virtual bool feedForward(CNeuronBaseOCL *NeuronOCL); ///< \brief Feed Forward method of calling kernel ::FeedForward().@param NeuronOCL Pointer to previos layer. ///\ingroup neuron_base_opt virtual bool updateInputWeights(CNeuronBaseOCL *NeuronOCL); ///< Method for updating weights.\details Calling one of kernels ::UpdateWeightsMomentum() or ::UpdateWeightsAdam() in depends of optimization type (#ENUM_OPTIMIZATION).@param NeuronOCL Pointer to previos layer. public: /** Constructor */CNeuronBaseOCL(void); /** Destructor */~CNeuronBaseOCL(void); virtual bool Init(uint numOutputs, uint myIndex, COpenCLMy *open_cl, uint numNeurons, ENUM_OPTIMIZATION optimization_type); ///< Method of initialization class.@param[in] numOutputs Number of connections to next layer.@param[in] myIndex Index of neuron in layer.@param[in] open_cl Pointer to #COpenCLMy object. #param[in] numNeurons Number of neurons in layer @param optimization_type Optimization type (#ENUM_OPTIMIZATION)@return Boolen result of operations. virtual void SetActivationFunction(ENUM_ACTIVATION value) { activation=value; } ///< Set the type of activation function (#ENUM_ACTIVATION) //--- virtual int getOutputIndex(void) { return Output.GetIndex(); } ///< Get index of output buffer @return Index virtual int getPrevOutIndex(void) { return PrevOutput.GetIndex(); } ///< Get index of previous iteration output buffer @return Index virtual int getGradientIndex(void) { return Gradient.GetIndex(); } ///< Get index of gradient buffer @return Index virtual int getWeightsIndex(void) { return Weights.GetIndex(); } ///< Get index of weights matrix buffer @return Index virtual int getDeltaWeightsIndex(void) { return DeltaWeights.GetIndex(); } ///< Get index of delta weights matrix buffer (SGD)@return Index virtual int getFirstMomentumIndex(void) { return FirstMomentum.GetIndex(); } ///< Get index of first momentum matrix buffer (Adam)@return Index virtual int getSecondMomentumIndex(void) { return SecondMomentum.GetIndex();} ///< Get index of Second momentum matrix buffer (Adam)@return Index //--- virtual int getOutputVal(double &values[]) { return Output.GetData(values); } ///< Get values of output buffer @param[out] values Array of data @return number of items virtual int getOutputVal(CArrayDouble *values) { return Output.GetData(values); } ///< Get values of output buffer @param[out] values Array of data @return number of items virtual int getPrevVal(double &values[]) { return PrevOutput.GetData(values); } ///< Get values of previous iteration output buffer @param[out] values Array of data @return number of items virtual int getGradient(double &values[]) { return Gradient.GetData(values); } ///< Get values of gradient buffer @param[out] values Array of data @return number of items virtual int getWeights(double &values[]) { return Weights.GetData(values); } ///< Get values of weights matrix buffer @param[out] values Array of data @return number of items virtual int Neurons(void) { return Output.Total(); } ///< Get number of neurons in layer @return Number of neurons virtual int Activation(void) { return (int)activation; } ///< Get type of activation function @return Type (#ENUM_ACTIVATION) virtual int getConnections(void) { return (CheckPointer(Weights)!=POINTER_INVALID ? Weights.Total()/(Gradient.Total()) : 0); } ///< Get number of connections 1 neuron to next layer @return Number of connections //--- virtual bool FeedForward(CObject *SourceObject); ///< Dispatch method for defining the subroutine for feed forward process. @param SourceObject Pointer to the previous layer. virtual bool calcHiddenGradients(CObject *TargetObject); ///< Dispatch method for defining the subroutine for transferring the gradient to the previous layer. @param TargetObject Pointer to the next layer. virtual bool UpdateInputWeights(CObject *SourceObject); ///< Dispatch method for defining the subroutine for updating weights.@param SourceObject Pointer to previos layer. ///\ingroup neuron_base_gr ///@{ virtual bool calcHiddenGradients(CNeuronBaseOCL *NeuronOCL); ///< Method to transfer gradient to previous layer by calling kernel ::CalcHiddenGradient(). @param NeuronOCL Pointer to next layer. virtual bool calcOutputGradients(CArrayDouble *Target); ///< Method of output gradients calculation by calling kernel ::CalcOutputGradient().@param Target target value ///@} //--- virtual bool Save(int const file_handle);///< Save method @param[in] file_handle handle of file @return logical result of operation virtual bool Load(int const file_handle);///< Load method @param[in] file_handle handle of file @return logical result of operation //--- virtual int Type(void) const { return defNeuronBaseOCL; }///< Identifier of class.@return Type of class };
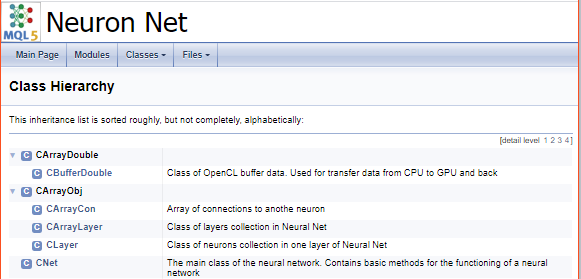
To finish working with the code, let us create a cover page. The "\mainpage" command is used to identify the cover page block. The command should be followed by the cover page title. Below, let us add the project description and create a list of references. The list items will be marked by character "-". To create links to earlier created groups, use the "\ref" command. When Doxygen generates documentation, pages of the class hierarchy (hierarchy.html) and of the files used (files.html) are generated. Add links to the specified pages to the list. The final code for the cover page is shown below.
///\mainpage NeuronNet /// Library for creating Neural network for use in MQL5 experts. /// - \ref const /// - \ref enums /// - \ref ObjectTypes /// - \ref group1 /// - [<b>Class Hierarchy</b>](hierarchy.html) /// - [<b>Files</b>](files.html)
The following page will be generated based on the above code.
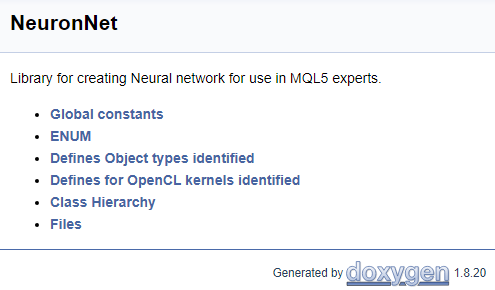
The full code of all comments is provided in the attachment.
5. Generating documentation
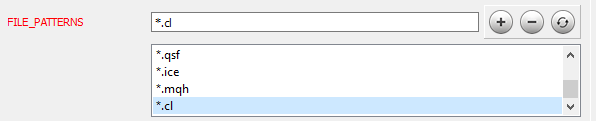
After completing working with the code, proceed to the next stage. Doxygen installation and setup is described in detail in article [9]. Let us consider the setting up of some program parameters. First, inform Doxygen which files it should work with: on the Expert tab, in the Input topic, add the necessary file masks to the FILE_PATTERNS parameter. In this case, I have added "*.mqh" and "*.cl".
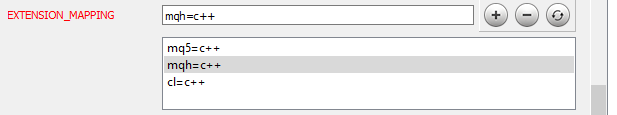
Now we need to inform Doxygen how to parse the added files. Go to the Project topic on the same Expert tab and edit the EXTENSION_MAPPING parameter as shown in the figure below.
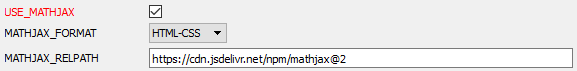
To enable Doxygen to generate mathematical formulas, activate the use of MathJax. To do this, activate the USE_MATHJAX parameter in the HTML topic of the Expert tab, as shown in the figure below.
After configuring the program, go to the Wizard tab and specify the name of the project, the path to the source files and the path for displaying the generated documentation (all these steps are shown in article [9]). Go to the Run tab and run the documentation generation program.
Once the program completes, you will receive a ready-to-use documentation. Some screenshots are shown below. The full documentation is provided in the attachment.
Conclusions
Documentation of developed programs is not the main task of the programmer. However, such documentation is essential when developing complex projects. It helps in tracking the implementation of tasks, in coordinating the work of a development team and simply provides a holistic view of the development. Documentation is a must when sharing knowledge.
The article describes a mechanism for documenting developments in the MQL5 language. It provides a detailed description of all steps of the mechanism. The results of the work performed are available in the attachment, so that everyone can evaluate them.
Hope my experience will be helpful.
References
- Neural networks made easy
- Neural networks made easy (Part 2): Network training and testing
- Neural networks made easy (Part 3): Convolutional networks
- Neural networks made easy (Part 4): Recurrent networks
- Neural networks made easy (Part 5): Multithreaded calculations in OpenCL
- Neural networks made easy (Part 6): Experimenting with the neural network learning rate
- Neural networks made easy (Part 7): Adaptive optimization methods
- Neural networks made easy (Part 8): Attention mechanisms
- Automatic creation of documentation for MQL5 programs
- Doxygen
Programs Used in the Article
| # | Name | Type | Description |
|---|---|---|---|
| 1 | NeuroNet.mqh | Class library | A library of classes for creating a neural network |
| 2 | NeuroNet.cl | Code Base | OpenCL program code library |
| 3 | html.zip | ZIP archive | Doxygen generated documentation archive |
| 4 | NN.chm | HTML Help | The converted HTML help file. |
| 5 | Doxyfile | Doxygen parameters file |
Translated from Russian by MetaQuotes Ltd.
Original article: https://www.mql5.com/ru/articles/8819
Warning: All rights to these materials are reserved by MetaQuotes Ltd. Copying or reprinting of these materials in whole or in part is prohibited.
This article was written by a user of the site and reflects their personal views. MetaQuotes Ltd is not responsible for the accuracy of the information presented, nor for any consequences resulting from the use of the solutions, strategies or recommendations described.
 The market and the physics of its global patterns
The market and the physics of its global patterns
 Developing a self-adapting algorithm (Part I): Finding a basic pattern
Developing a self-adapting algorithm (Part I): Finding a basic pattern
 Finding seasonal patterns in the forex market using the CatBoost algorithm
Finding seasonal patterns in the forex market using the CatBoost algorithm
 Neural networks made easy (Part 8): Attention mechanisms
Neural networks made easy (Part 8): Attention mechanisms
- Free trading apps
- Over 8,000 signals for copying
- Economic news for exploring financial markets
You agree to website policy and terms of use
Not really. We check the values of the target, just like in hidden layers we add outpuVal to the gradient to get the target and check its value. The point is that sigmoid has a limited range of results: logistic function from 0 to 1, tanh - from -1 to 1. If we penalise the neuron for deviation and increase the weighting factor indefinitely, we will come to weight overflow. After all, if we came to a neuron's value equal to 1, and the subsequent layer transmitting an error says that we should increase the value to 1.5. The neuron will obediently increase the weights at each iteration, and the activation function will cut off the values at the level of 1. Therefore, I limit the values of the target to the ranges of acceptable values of the activation function. And I leave the adjustment outside the range to the weights of the subsequent layer.
I think I've got it. But I'm still wondering if this is the right approach, an example like this:
if the network makes a mistake by giving 0 when it's really 1. From the last layer then the gradient weighted on the previous layer comes (most likely, as I understand) positive and can be more than 1, let's say 1,6.
Suppose there is a neuron in the previous layer that produced +0.6, i.e. it produced the correct value - its weight should increase in plus. And with this normalisation we cut the change in its weight.
The result is norm(1,6)=1. 1-0,6=0,4, and if we normalise it as I suggested, it will be 1. In this case, we inhibit the amplification of the correct neuron.
What do you think?
About infinite increase of weights, something like I heard that it happens in case of "bad error function", when there are a lot of local minima and no expressed global, or the function is not convex, something like that, I'm not a super expert, I just believe that you can and should fight with infinite weights and other methods.
I'm asking for an experiment to test both variants. If I think of how to formulate the test )
When saving-reading a network from a file, there is an error on the transformers layer.
in the method
bool CLayer::CreateElement(int index)
in the line
int type=FileReadInteger(iFileHandle);
reads 0 and switch goes to default false.
(Apparently, there is a write asynchrony).
If it has already been fixed, please give me a hint for speed, or send me the file.
Well that the same thing is not to fix twice and just do not want to make a lot of changes to the library.
For work in the tester made changes, and then you forget when a new article comes out that you need to transfer to the new version of your edits).
When saving-reading a network from a file, there is an error on the transformers layer.
in the method
bool CLayer::CreateElement(int index)
in the string
int type=FileReadInteger(iFileHandle);
reads 0 and switch goes to default false.
(Apparently, there is a write asynchrony)
If it is already fixed, please give me a hint for speed, or send me the file.
Well that one and the same not to fix twice and just do not want to make a lot of changes to the library.
For work in the tester made changes, and then you forget when a new article comes out that you need to transfer your edits to the new version).
Good day,
Now a new article is being checked by a moderator. This error is fixed there.
Good afternoon,
There is a new article now being checked by a moderator. This error has been corrected there.
Great! Let's wait.