
Population optimization algorithms: Saplings Sowing and Growing up (SSG)
Contents:
1. Introduction
2. Algorithm
3. Test results
1. Introduction
All living organisms in nature are subject to certain laws. This helps them to survive in changing environmental conditions. There are various options for the adaptability of plants to the environment. Some of them are able to tolerate seasonal changes, others can adapt to lack of moisture, high or low temperatures and the absence of pollinators. One of the most stable organisms among plants are trees, some of which are able to live for more than 50 thousand years forming colonies.
Nature is an inexhaustible field of inspiration for many effective ideas in the development and invention of computational methods. In fact, evolutionary computing is a projection of evolution into computer simulations. There are many optimization methods inspired by processes occurring in nature, such as evolutionary computation, artificial immunology, population methods and others. SSG is basically defined as iterative generation and combination processes working with a garden of potential solutions called seedlings. The Saplings Sowing and Growing (SSG) algorithm was proposed by A. Karci with co-authors in 2002. The algorithm is inspired by the evolution of growing trees and models the growth and branching of trees.
2. Algorithm
The algorithm is one of the few that does not have a clear description by the authors (only general provisions and ideas are provided). The algorithm operators presented by the authors are also not ready-made instructions for the algorithmic implementation of the program. There are no clear instructions about child and parent trees and their interaction. There are no requirements for the order, in which operators are executed, and any user can change their order to obtain a better seedling.
In a broad sense, SSG is not an optimization algorithm, it is a general set of rules that is designed to complement other algorithms to improve the quality of optimization. In other words, SSG is an add-on for any evolutionary population algorithms, so I have room for imagination and the opportunity to experiment with a specific implementation of the optimization algorithm. I applied some of my own thoughts and experience while writing previous algorithms and used them to work with SSG. The results of the experiments are presented for the reader's judgment below.
To start understanding the algorithm, we need to think of a tree as an optimization agent. A tree is a solution to an optimization problem, where each branch is an optimized parameter of the problem. An abstract and artistic illustration of child and parent trees is provided in Figure 1. The tree trunk is a set of parameters to be optimized. Each branch is a separate optimized parameter, where the length of the branch is limited by the allowable range of values of the corresponding parameter. The direction of the branches does not matter and is only shown in the figure to highlight their difference.
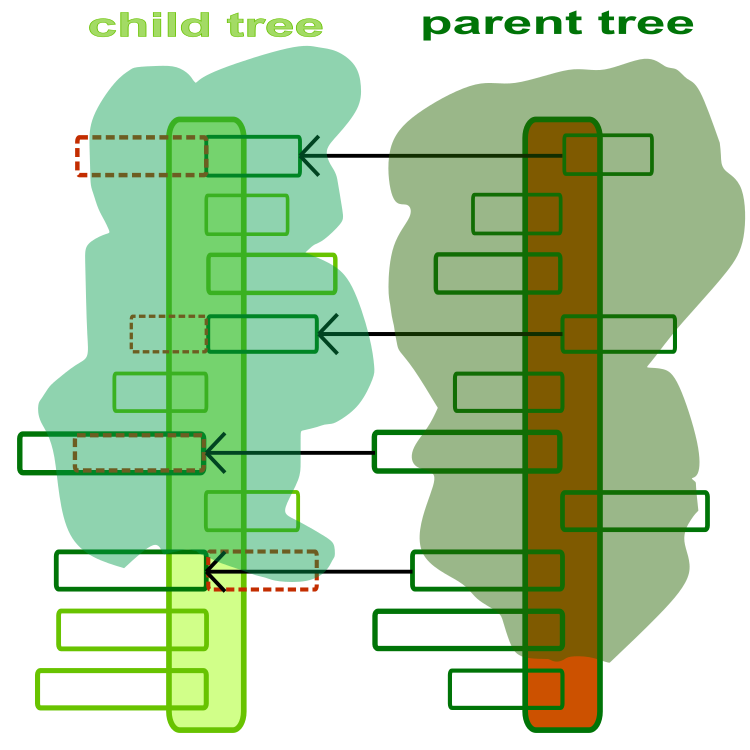
Figure 1. Child and parent tree. The dotted line indicates the child branches replaced by the parent ones
Thus, the tree branches are the coordinates of the trees in the search space.
The SSG algorithm consists of variation operators that generate new solutions - candidates for the common pool of solutions. The main variation operators arecrossing, branching andvaccination. Planting seedlings should be carried out at an equidistant distance in each direction from each other (west, east, north, south), and this is the initial stage of the method. When the coordinates (optimized parameters) are much more than three, then "uniform" planting is a simple random distribution of seedlings over the search space. Then the seedlings grow up, interbreed, branch and the vaccination process takes place.
Let's consider the steps and operators of the SSG algorithm:
1) Planting seedlings.
The search space can be thought of as a garden of seedlings, and hence all seedlings must be evenly distributed throughout the garden. When sowing seedlings, a farmer simply sows them at an equal distance from each other so that the seedlings grow faster without interfering with each other. In order to solve the problem by simulating the cultivation of seedlings, the arbitrary solutions to be generated initially must be evenly distributed in the valid search space.
When there are two or three coordinates, then there is no problem to evenly distribute seedlings, but when there are much more than three coordinates, it is easier to use a random distribution. In practice, with a small number of coordinates, there is no need to take care of the uniform distribution of seedlings, since the task is not a big problem and it is known that the solution will be obtained with high accuracy. Therefore, regardless of the number of coordinates in the algorithm, we will use a random distribution of seedlings in the garden.
2) Growing seedlings (trees), three operators executed in sequence.
2.1) Crossing.
The purpose of the "crossing" operator is to create a new seedling from currently existing seedlings by exchanging information between them. Crossing is essentially a transplant of a copy of a branch from the parent tree to the child one (Figure 1). For each pair of seedlings, a different crossover coefficient is used, which is the probability of crossover. The probability of crossing depends on the distance between seedlings in a pair, the greater the distance, the lower the probability of crossing. The crossing operator is a very important method of the algorithm that provides combinatorial mechanisms. Combining information between agents can significantly improve the overall quality of the optimization algorithm.
2.2) Branching.
The operator models the growth of branches. The growth can be either positive (elongation) or negative (drying). "In order to grow a branch at any point in the seedling's body, there will be no nearby branch that has previously sprouted there". This an approximate description of the operator by the authors of the algorithm. In fact, this process is simpler and clearer than it might seem at first glance, and is a modification of the existing branches of the child tree (the specific method of modification is not specified).
Modification of each individual branch is the more likely, the more iterations have passed between the current iteration and the one on which the last modification of the branch was made. My experiments showed the inefficiency of this operator. Besides, there are no direct indications of the use of the modification method, and I took the initiative in this matter and applied the change in the length of the branch according to the Levy flight distribution law. The modification will be performed with the probability and intensity specified as external parameters of the algorithm.
2.3) Vaccination.
The vaccination process takes place between two different seedlings in case of seedling similarity. The similarity of seedlings affects the success of the vaccination process and is proportional to the arithmetic mean of the weighted distance. The operator is similar to the crossing operator and consists in the exchange of branches, providing the algorithm with an additional way to combine information between agents. The operator will be highlighted in the article, but this operator is commented out in the source codes and the test results are presented without its participation, since vaccination worsens the results.
3) Calculating fitness of trees.
4) Planting new seedlings in the garden.
The seedlings obtained with the help of the crossing, branching and vaccination operators are temporary solutions (daughter garden). At this stage, it is necessary to select the n best seedlings (an external parameter of the algorithm) and place them in the garden replacing the n worst trees in the garden. It should be noted that the replacement with seedlings will occur in any case, even if they are worse than the worst trees in the garden.
This is a good time to look at the code of the growing tree algorithm, bringing us steadily closer to the exciting climax of this study - the review of test results.
So, it is convenient to represent each tree as a Garden structure, which will serve as the basis for creating a flowering garden. There is nothing simpler in this algorithm than the "tree" entity, which needs only two components: the coordinates with [] and the fitness value f.
//—————————————————————————————————————————————————————————————————————————————— struct S_Garden { double c []; //coordinates double f; //fitness }; //——————————————————————————————————————————————————————————————————————————————
The C_AO_SSG class of the SG algorithm is nothing special. Everything here is very familiar to us from the algorithms considered earlier. In the class, we will declare members and methods for operating on the parent and child gardens. A temporary garden is meant for the sorting method to function, because we need to sort both the child and parent gardens. Let's declare an array of the best coordinates of the solution as a whole and the best fitness value, as well as constant variables for storing the external parameters of the algorithm.
//—————————————————————————————————————————————————————————————————————————————— class C_AO_SSG { //============================================================================ public: double rangeMax []; //maximum search range public: double rangeMin []; //manimum search range public: double rangeStep []; //step search public: S_Garden pGarden []; //parent's garden public: S_Garden cGarden []; //child's garden public: S_Garden gardenT []; //temp garden public: double cB []; //best coordinates public: double fB; //fitness of the best coordinates public: void Init (const int coordinatesP, //Number of coordinates const int numberTreesP, //Number of trees const double seedlingsReplacementP, //Seedlings replacement const double probabMatingOperatorP, //Probability mating operator const double probabBranchOperatorP, //Probability branching operator const double powerBranchOperatorP); //Power branching operator public: void Sowing (int iter); public: void Germination (); //============================================================================ private: void Sorting (S_Garden &garden []); private: double SeInDiSp (double In, double InMin, double InMax, double Step); private: double RNDfromCI (double Min, double Max); private: double Scale (double In, double InMIN, double InMAX, double OutMIN, double OutMAX, bool Revers); private: double vec []; //Vector private: int ind []; private: double val []; private: double r; private: bool sowing; //Sowing private: int coordinates; //Coordinates number private: int numberTrees; //Number of trees private: int seedlingsReplacement; //Seedlings replacement private: double probabMatingOperator; //Probability mating operator private: double probabBranchOperator; //Probability branching operator private: double powerBranchOperator; //Power branching operator }; //——————————————————————————————————————————————————————————————————————————————
In the Init () initialization method, allocate memory for arrays and assign values to constant parameters. Since the seedlingsReplacementP parameter is set in fractions of the garden size (from 0.0 to 1.0), which is responsible for the number of child seedlings to plant in the parent garden, it should be converted into an integer representation of the garden size. Let's reset the initial garden seedling flag and initialize the global decision variable with the smallest possible double value.
//—————————————————————————————————————————————————————————————————————————————— void C_AO_SSG::Init (const int coordinatesP, //Number of coordinates const int numberTreesP, //Number of trees const double seedlingsReplacementP, //Seedlings replacement const double probabMatingOperatorP, //Probability mating operator const double probabBranchOperatorP, //Probability branching operator const double powerBranchOperatorP) //Power branching operator { MathSrand (GetTickCount ()); sowing = false; fB = -DBL_MAX; coordinates = coordinatesP; numberTrees = numberTreesP; if (seedlingsReplacementP >= 1.0) { seedlingsReplacement = numberTreesP; } else { if (seedlingsReplacementP <= 0.0) { seedlingsReplacement = 1; } else seedlingsReplacement = int(numberTreesP * seedlingsReplacementP); } probabMatingOperator = probabMatingOperatorP; probabBranchOperator = probabBranchOperatorP; powerBranchOperator = powerBranchOperatorP; ArrayResize (rangeMax, coordinates); ArrayResize (rangeMin, coordinates); ArrayResize (rangeStep, coordinates); ArrayResize (vec, coordinates); ArrayResize (cB, coordinates); ArrayResize (pGarden, numberTrees); ArrayResize (cGarden, numberTrees); ArrayResize (gardenT, numberTrees); ArrayResize (ind, numberTrees); ArrayResize (val, numberTrees); for (int i = 0; i < numberTrees; i++) { ArrayResize (pGarden [i].c, coordinates); ArrayResize (cGarden [i].c, coordinates); ArrayResize (gardenT [i].c, coordinates); cGarden [i].f = -DBL_MAX; } } //——————————————————————————————————————————————————————————————————————————————
The first public method called on every iteration, Sowing() is seeding. At the first iteration, when the algorithm is just running, we will scatter seedlings around the garden (search space) randomly with a uniform distribution. Here we see that the coordinates are generated randomly in the allowable range between the min and max of the optimized parameters, we check for an exit from the allowable range, then we bring the coordinate values in accordance with the optimization step.
At this stage, the adaptability of seedlings is minimal. We set the vec[] vector. We will need it to scale the increments of the branches in the branching operator and also calculate the maximum possible Euclidean distance r in the search space. It will be useful in the crossing operator to determine the probability depending on the distance between pairs of trees.
//the first planting of trees------------------------------------------------- if (!sowing) { fB = -DBL_MAX; r = 0.0; for (int t = 0; t < numberTrees; t++) { for (int c = 0; c < coordinates; c++) { cGarden [t].c [c] = RNDfromCI (rangeMin [c], rangeMax [c]); cGarden [t].c [c] = SeInDiSp (cGarden [t].c [c], rangeMin [c], rangeMax [c], rangeStep [c]); } cGarden [t].f = -DBL_MAX; } for (int c = 0; c < coordinates; c++) { vec [c] = rangeMax [c] - rangeMin [c]; r += vec [c] * vec [c]; } r = sqrt (r); return; }
Next, in the Sowing() method, the crossing, branching and vaccination operators are executed at the second and subsequent iterations. The operators are executed sequentially in the main loop:
//tree growth----------------------------------------------------------------- int child, parent; double rnd; double ed; //euclidean distance double eM; double u; double temp; for (int t = 0; t < numberTrees; t++)
When executing operators, the concepts of "child" and "parent" are very arbitrary. In fact, they are just sources of basic information for creating a new seedling. The new seedling copies all branches (as you might remember, branches are the parameters to be optimized) from the child tree and receives new ones from the parent.
For each new seedling, two trees are individually selected from the garden, a child and a parent one at random, equally likely for all trees in the garden. In other words, all trees can take part in the creation of a new seedling with equal probability and regardless of their fitness. Thus, 'child' and 'parent' are simply the indices of the original two trees in the parent garden array.
ed = 0.0; rnd = RNDfromCI (0.0, numberTrees - 1); child = (int)MathRound (rnd); if (child < 0) child = 0; if (child > numberTrees - 1) child = numberTrees - 1; rnd = RNDfromCI (0.0, numberTrees - 1); parent = (int)MathRound (rnd); if (parent < 0) parent = 0; if (parent > numberTrees - 1) parent = numberTrees - 1; if (child == parent) parent++; if (parent > numberTrees - 1) parent = 0; ArrayCopy (cGarden [t].c, pGarden [child].c, 0, 0, WHOLE_ARRAY);
The first operator is crossing, or conjugation (mating). To execute the crossing operator on a seedling with index t, it is necessary to calculate the Euclidean space between the child and parent trees with the 'child' and 'parent' indices:
//mating operator----------------------------------------------------------- for (int c = 0; c < coordinates; c++) { temp = pGarden [child].c [c] - pGarden [parent].c [c]; ed += temp * temp; } ed = sqrt (ed);
The Euclidean distance is involved in the equation for calculating the crossing probability through normalization to the maximum possible Euclidean distance r in the search space:
eM = 1.0 - (ed / r); Generate a random number in the range [0.0;1.0]. If the resulting number falls within the calculated probability eM, then we perform crossing - copying branches from the parent tree with the probabMatingOperator probability for each branch. If the eM probability is not satisfied, then the seedling will proceed to the execution of the next statement with all the original branches of the child tree.
rnd = RNDfromCI (0.0, 1.0); if (rnd <= eM) { for (int c = 0; c < coordinates; c++) { rnd = RNDfromCI (0.0, 1.0); if (rnd <= probabMatingOperator) cGarden [t].c [c] = pGarden [parent].c [c]; } }
Next, the branch operator is executed. Branching provides a variation of branches - lengthening and shortening. This is the main operator that creates branches of a new length. The remaining operators perform only a combinatorial role and do not create new unique branches. For each branch, we generate a random number in the range [0.0;1.0] and if the probability of probabBranchOperator is fulfilled, then we perform branching by changing the length of the branch according to the Levy flights distribution law consideredhere.
As you can see, not all branches of the seedling will be changed. They will be changed regardless of whether the branch was copied from the parent tree in the previous statement, or it is the original child branch. After modifying the branch, we check for out-of-range values and bring it into line with the optimization step.
//branching operator-------------------------------------------------------- for (int c = 0; c < coordinates; c++) { rnd = RNDfromCI (0.0, 1.0); if (rnd < probabBranchOperator) { double r1 = RNDfromCI (0.0, 1.0); r1 = r1 > 0.5 ? 1.0 : -1.0; double r2 = RNDfromCI (1.0, 20.0); cGarden [t].c [c] = cGarden [t].c [c] + r1 * vec [c] * powerBranchOperator * pow (r2, -2.0); cGarden [t].c [c] = SeInDiSp (cGarden [t].c [c], rangeMin [c], rangeMax [c], rangeStep [c]); } }
The third operator is vaccination. The vaccination operator was conceived by the authors, apparently, as a combinatorial operator, and should be performed to copy branches from the parent tree in the case when the branches of the child and parent tree differed by more than the average difference across all branches of the pair. It sounds very complicated, but there is little use for this operator, and therefore this operator is commented out in the source files. In this case, I cannot act as an expert, and therefore I admit that I may not have correct understanding of this operator.
//vaccinating operator------------------------------------------------------ u = 0.0; for (int c = 0; c < coordinates; c++) { u += fabs (cGarden [t].c [c] - pGarden [parent].c [c]) / vec [c]; } u /= coordinates; for (int c = 0; c < coordinates; c++) { if (fabs (cGarden [t].c [c] - pGarden [parent].c [c]) / vec [c] >= u) { cGarden [t].c [c] = pGarden [parent].c [c]; } }
The time has come for the last, but not the least important, operation of the algorithm - germination. We execute the second public method Germination () that is mandatory for execution at each iteration.
If the first iteration is coming to an end (we will certainly know this by the 'sowing' flag), then this means that the parent garden is empty. We plant all the seedlings from the child garden in the parent garden simply by copying all the young trees one by one. After that, sort the parent garden by fitness value using the Sorting() method. If the trees are sorted, then the tree at index 0 will have the best fitness, and we can update the best global solution if it is not the best already.
if (!sowing) { for (int t = 0; t < numberTrees; t++) pGarden [t] = cGarden [t]; Sorting (pGarden); if (pGarden [0].f > fB) { fB = pGarden [0].f; ArrayCopy (cB, pGarden [0].c, 0, 0, WHOLE_ARRAY); } sowing = true; return; }
Further in the code, we will only get to the second and subsequent iterations in the algorithm, since the 'sowing' flag has been prudently set to 'true'. On the second and subsequent iterations, the child garden must be sorted before the seedlings are transferred to the parent garden and the worst trees are replaced. Let's check if the global solution has improved, and then copy the best n seedlings to the end of the parent garden.
In conclusion, it remains only to sort the parent garden. The new members of the garden society will be able to replace the trees of older generations to please us with flowers results of solving optimization problems.
//planting some part from all child trees------------------------------------- Sorting (cGarden); if (cGarden [0].f > fB) { fB = cGarden [0].f; ArrayCopy (cB, cGarden [0].c, 0, 0, WHOLE_ARRAY); } for (int t = 0; t < seedlingsReplacement; t++) pGarden [numberTrees - seedlingsReplacement + t] = cGarden [t]; Sorting (pGarden);
3. Test results
SSG test stand results:
2023.03.09 12:50:37.207 Test_AO_SSG (GBPUSD,M1) C_AO_SSG:50;0.3;0.5;0.4;0.1
2023.03.09 12:50:37.207 Test_AO_SSG (GBPUSD,M1) =============================
2023.03.09 12:50:45.954 Test_AO_SSG (GBPUSD,M1) 5 Rastrigin's; Func runs 10000 result: 80.7052793572295
2023.03.09 12:50:45.954 Test_AO_SSG (GBPUSD,M1) Score: 0.99998
2023.03.09 12:50:59.290 Test_AO_SSG (GBPUSD,M1) 25 Rastrigin's; Func runs 10000 result: 77.3972262608643
2023.03.09 12:50:59.290 Test_AO_SSG (GBPUSD,M1) Score: 0.95900
2023.03.09 12:52:25.368 Test_AO_SSG (GBPUSD,M1) 500 Rastrigin's; Func runs 10000 result: 52.24518909141432
2023.03.09 12:52:25.368 Test_AO_SSG (GBPUSD,M1) Score: 0.64735
2023.03.09 12:52:25.368 Test_AO_SSG (GBPUSD,M1) =============================
2023.03.09 12:52:31.419 Test_AO_SSG (GBPUSD,M1) 5 Forest's; Func runs 10000 result: 1.331178589711503
2023.03.09 12:52:31.419 Test_AO_SSG (GBPUSD,M1) Score: 0.75298
2023.03.09 12:52:42.575 Test_AO_SSG (GBPUSD,M1) 25 Forest's; Func runs 10000 result: 1.019329018074209
2023.03.09 12:52:42.575 Test_AO_SSG (GBPUSD,M1) Score: 0.57658
2023.03.09 12:53:48.922 Test_AO_SSG (GBPUSD,M1) 500 Forest's; Func runs 10000 result: 0.25410121872226443
2023.03.09 12:53:48.922 Test_AO_SSG (GBPUSD,M1) Score: 0.14373
2023.03.09 12:53:48.922 Test_AO_SSG (GBPUSD,M1) =============================
2023.03.09 12:53:57.201 Test_AO_SSG (GBPUSD,M1) 5 Megacity's; Func runs 10000 result: 6.4
2023.03.09 12:53:57.201 Test_AO_SSG (GBPUSD,M1) Score: 0.53333
2023.03.09 12:54:08.004 Test_AO_SSG (GBPUSD,M1) 25 Megacity's; Func runs 10000 result: 4.504
2023.03.09 12:54:08.004 Test_AO_SSG (GBPUSD,M1) Score: 0.37533
2023.03.09 12:56:07.061 Test_AO_SSG (GBPUSD,M1) 500 Megacity's; Func runs 10000 result: 1.2336
2023.03.09 12:56:07.061 Test_AO_SSG (GBPUSD,M1) Score: 0.10280
2023.03.09 12:56:07.061 Test_AO_SSG (GBPUSD,M1) =============================
2023.03.09 12:56:07.061 Test_AO_SSG (GBPUSD,M1) All score: 5.09109
SSG does not have too many parameters, although this is a consequence of tweaking the algorithm (original SSG has even fewer parameters). However, all the parameters deserve exceptional attention. The following parameters were used in the tests. As you might remember, in each article I provide the best algorithm parameters yielding the highest possible result in the tests.
C_AO_SSG:50;0.3;0.5;0.4;0.1
input int NumberTrees_P = 50; - number of trees in the garden. I did not experiment with this parameter leaving the default value (the default in my experiments). In case of the value of 100, the algorithm produces yields even better aggregate results but the ability to scale decreases due to a decrease in the number of allowable iterations by garden given the size of the garden. A garden with a large number of branches does not have time to evolve.
input double SeedlingsReplacement_P = 0.3; - proportion of seedlings from the daughter garden to be transferred to the parent one.
input double ProbabMatingOperator_P = 0.5; - probability of crossing (copying branches from the parent tree) if the probability taking into account the distance between a pair of trees is fulfilled.
input double ProbabBranchOperator_P = 0.4; - probability of branching (growth/shrinkage of branches). This is an important parameter that significantly affects the performance of the algorithm (generally, all parameters are important).
input double PowerBranchOperator_P = 0.1; - branching strength. This is a scaling factor in the dimension of the parameters being optimized, 1.0 or more will mean the growth of branches to the boundaries of the garden, 0.0 - no growth of branches, that is, new branch sizes will not arise and the algorithm degenerates into a simple combinatorics tool (which is probably a great idea for using SSG, however more research is required here).
If you pay attention to the animation of the SSE algorithm on test functions, you will notice the absence of any patterns of tree movement in the garden, only some "clumping" in local extrema is noticeable. However, the high quality of convergence is immediately noticeable when compared with the previously considered algorithms. The stable reproducibility of the results is also notable.
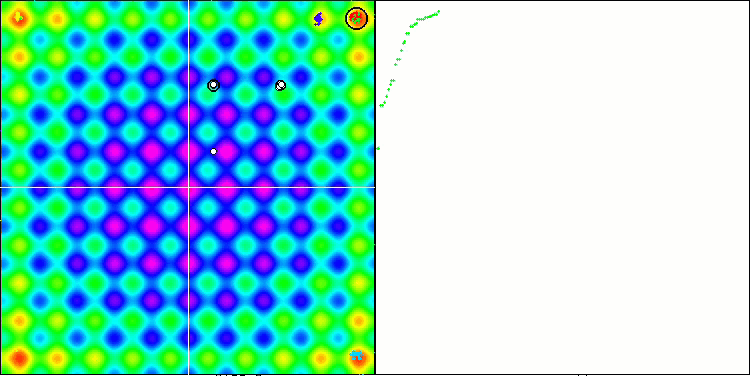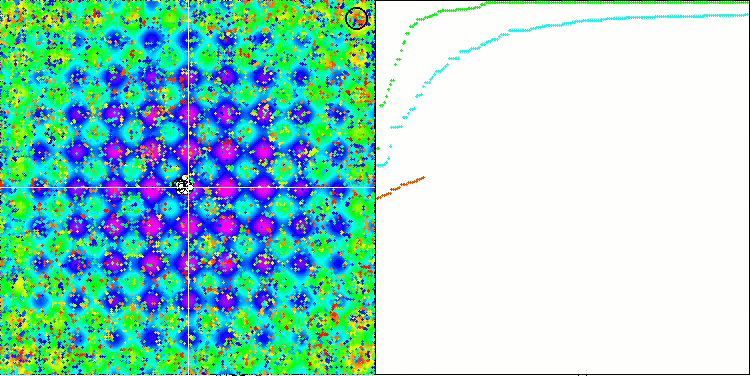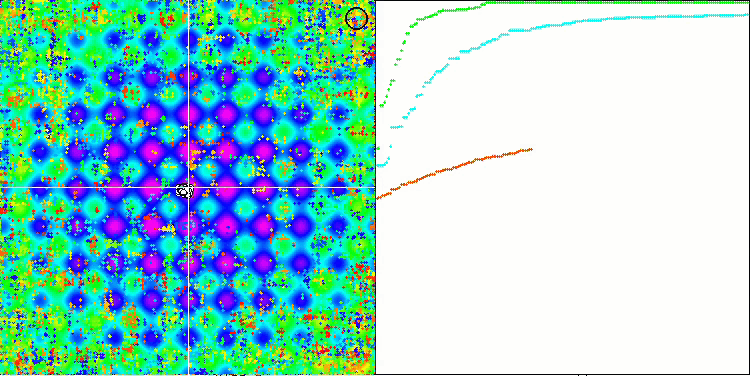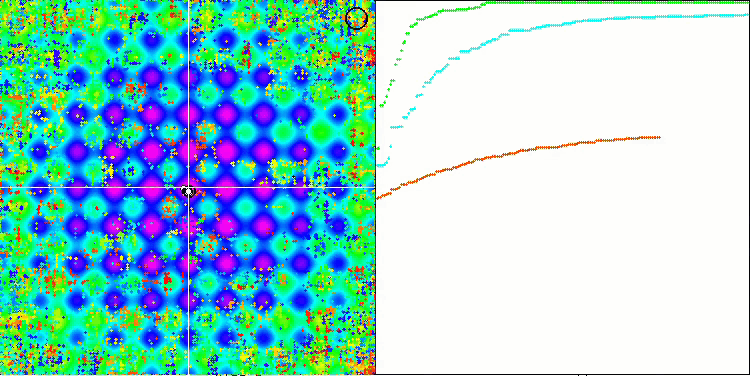
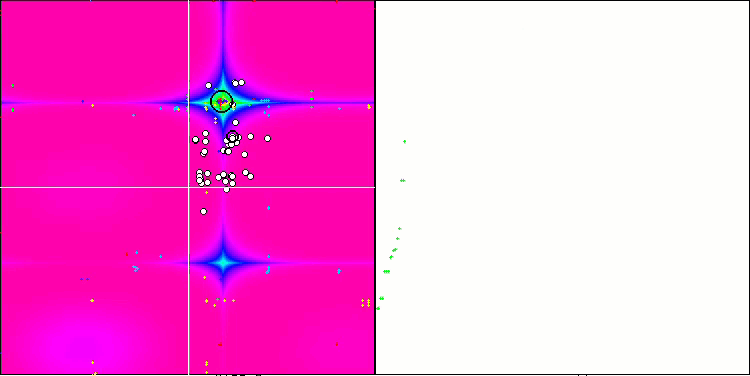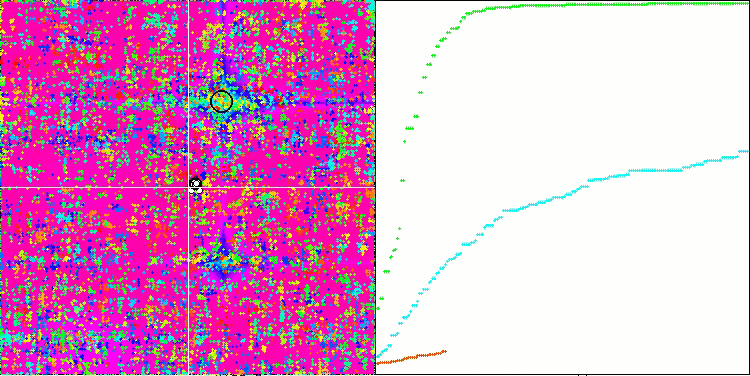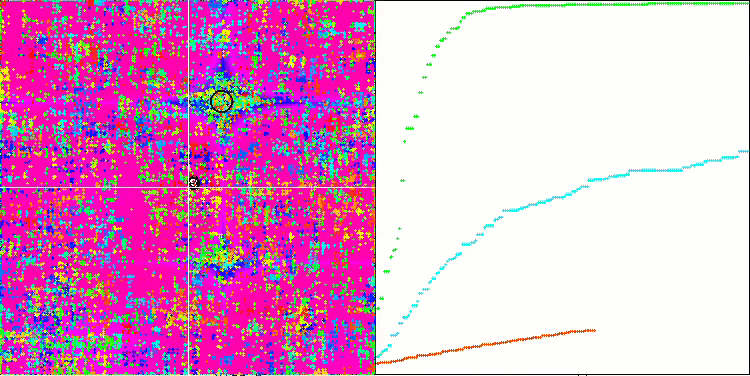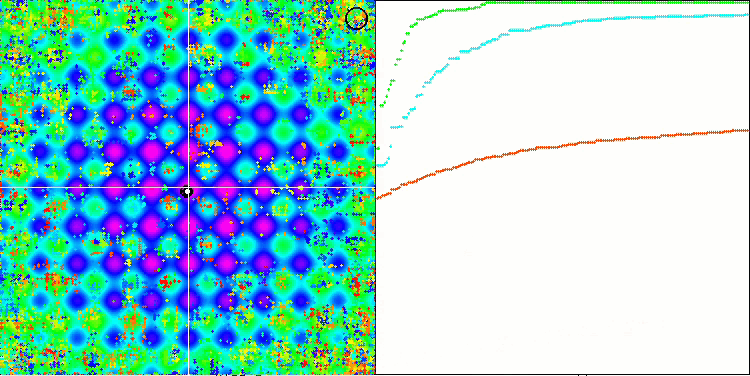
SSG on the Rastrigin test function.
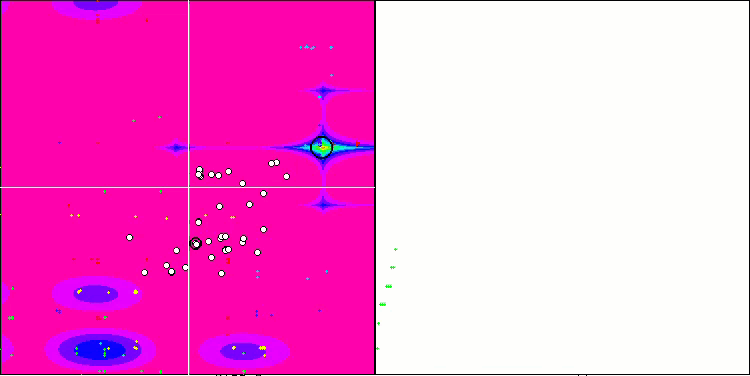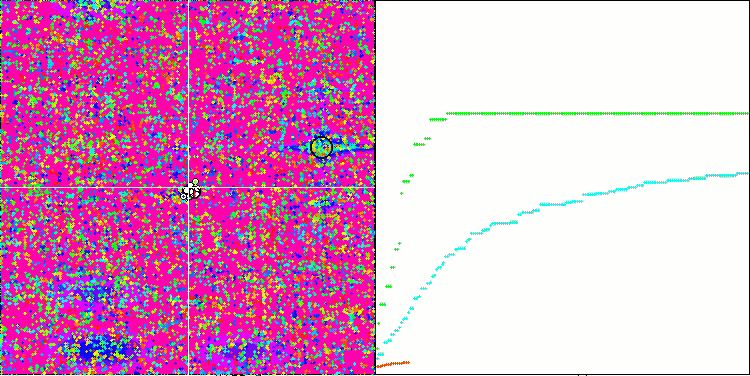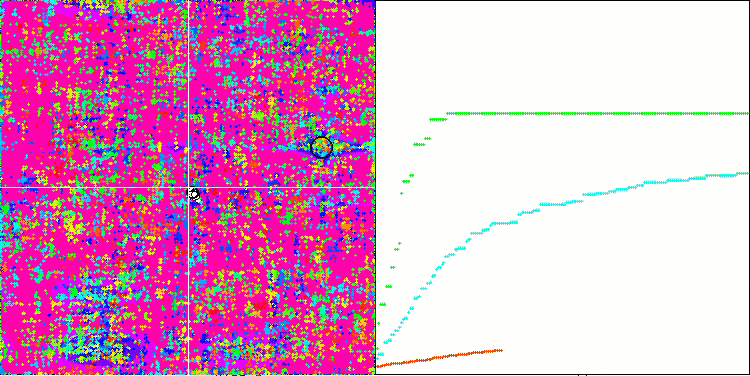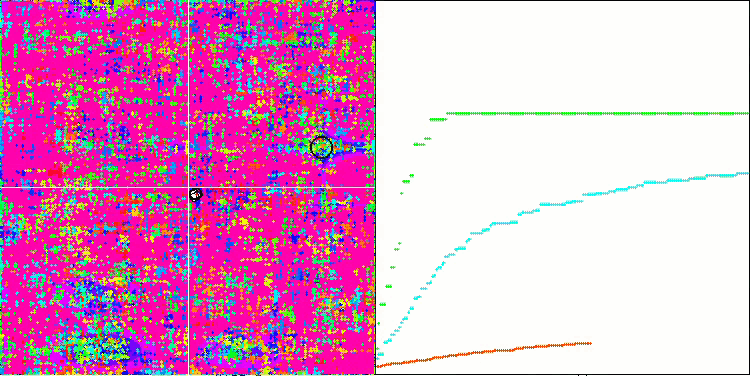
SSG on the Forest test function.
SSG on the Megacity test function.
Let's move on to discussing the results of the SSG algorithm. There is definitely something to talk about. The algorithm has triumphantly burst into the first place of the rating leaving rivals behind! The algorithm does not use fitness knowledge directly to modify decision trees. The fitness is only needed to sort the child garden and the parent garden, so it is all the more surprising that the algorithm has been able to show such remarkable results in testing.
This is the same as if the team of the blind beat the team of the sighted in orienteering. The algorithm was ahead of the participants in the table in four out of six tests, while remaining not far behind in the tests where it was not a leader. SSG showed the most impressive lead over rivals on the Megacity discrete function outperforming the closest competitor HS by almost 60%! This demonstrates the excellent scalability of the algorithm. The best scalability result was also observed on the "sharp" Forest test function. SSG outperformed the best competitor in this test (ACOm) by almost 30%.
| AO | Description | Rastrigin | Rastrigin final | Forest | Forest final | Megacity (discrete) | Megacity final | Final result | ||||||
| 10 params (5 F) | 50 params (25 F) | 1000 params (500 F) | 10 params (5 F) | 50 params (25 F) | 1000 params (500 F) | 10 params (5 F) | 50 params (25 F) | 1000 params (500 F) | ||||||
| SSG | saplings sowing and growing | 1.00000 | 1.00000 | 0.65914 | 2.65914 | 0.70823 | 0.94522 | 1.00000 | 2.65345 | 0.71532 | 0.85412 | 1.00000 | 2.56944 | 100.000 |
| HS | harmony search | 0.99676 | 0.95282 | 0.57048 | 2.52006 | 1.00000 | 0.98931 | 0.44806 | 2.43736 | 1.00000 | 1.00000 | 0.41537 | 2.41537 | 93.370 |
| ACOm | ant colony optimization M | 0.34611 | 0.17985 | 0.20182 | 0.72778 | 0.85966 | 1.00000 | 0.77362 | 2.63327 | 1.00000 | 0.88484 | 0.05606 | 1.94090 | 66.407 |
| IWO | invasive weed optimization | 0.95828 | 0.67083 | 0.35295 | 1.98207 | 0.68718 | 0.46349 | 0.31773 | 1.46840 | 0.75912 | 0.39732 | 0.33289 | 1.48933 | 61.691 |
| COAm | cuckoo optimization algorithm M | 0.92400 | 0.46794 | 0.30792 | 1.69987 | 0.55451 | 0.34034 | 0.16526 | 1.06012 | 0.67153 | 0.30326 | 0.17083 | 1.14561 | 48.226 |
| FAm | firefly algorithm M | 0.59825 | 0.33980 | 0.20290 | 1.14095 | 0.47632 | 0.42299 | 0.49790 | 1.39721 | 0.21167 | 0.25143 | 0.35258 | 0.81568 | 41.042 |
| ABC | artificial bee colony | 0.78170 | 0.32715 | 0.24656 | 1.35541 | 0.50591 | 0.21455 | 0.13344 | 0.85390 | 0.47444 | 0.23609 | 0.13926 | 0.84979 | 37.204 |
| BA | bat algorithm | 0.40526 | 0.63761 | 1.00000 | 2.04287 | 0.15275 | 0.17477 | 0.25989 | 0.58741 | 0.15329 | 0.06334 | 0.17371 | 0.39034 | 36.703 |
| GSA | gravitational search algorithm | 0.70167 | 0.45217 | 0.00000 | 1.15384 | 0.26854 | 0.36416 | 0.33204 | 0.96475 | 0.51095 | 0.32436 | 0.00000 | 0.83531 | 35.834 |
| BFO | bacterial foraging optimization | 0.67203 | 0.30963 | 0.13988 | 1.12154 | 0.35462 | 0.26623 | 0.20652 | 0.82736 | 0.42336 | 0.30519 | 0.18932 | 0.91786 | 34.700 |
| MA | monkey algorithm | 0.33192 | 0.33451 | 0.17340 | 0.83983 | 0.03684 | 0.07891 | 0.08932 | 0.20508 | 0.05838 | 0.00383 | 0.10720 | 0.16941 | 13.185 |
| FSS | fish school search | 0.46812 | 0.25337 | 0.13383 | 0.85532 | 0.06711 | 0.05013 | 0.06516 | 0.18240 | 0.00000 | 0.00959 | 0.08283 | 0.09242 | 12.089 |
| PSO | particle swarm optimisation | 0.20449 | 0.08200 | 0.08478 | 0.37127 | 0.13192 | 0.10486 | 0.21738 | 0.45415 | 0.08028 | 0.02110 | 0.01957 | 0.12095 | 9.696 |
| RND | random | 0.16826 | 0.09743 | 0.09495 | 0.36065 | 0.07413 | 0.04810 | 0.04715 | 0.16938 | 0.00000 | 0.00000 | 0.04922 | 0.04922 | 4.916 |
| GWO | grey wolf optimizer | 0.00000 | 0.00000 | 0.02672 | 0.02672 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.18977 | 0.03645 | 0.02557 | 0.25179 | 1.000 |
Summary
SSG features: no requirements for differentiability and continuity of the optimized function, no restrictions on the type of applied representation and encodings. The algorithm does not use fitness information of individual agents and the best solution as a whole. Thanks to these features, SSG can be easily applied to various optimization problems, including very complex ones. SSG can be definitely recommended for use in traders' problems and machine learning. At the time of this writing, the Saplings Sowing and Growing up algorithm is the undisputed leader in the quality of convergence, stability of results and scalability.
Fig. 2 shows the test results of the algorithm
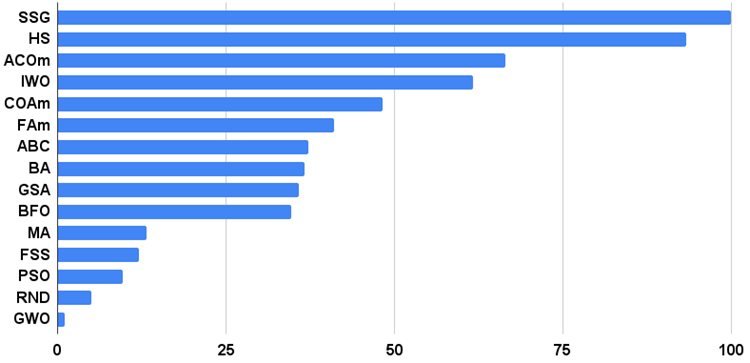
Figure 2. Histogram of the test results of the algorithms
SSG algorithms pros and cons:
Pros:
1. Easy implementation.
2. Excellent convergence on all types of functions with no exception.
3. Impressive scalability.
Cons:
1. Lots of customization options, although they are intuitively clear.
Each article features an archive that contains updated current versions of the algorithm codes for all previous articles. The article is based on the accumulated experience of the author and represents his personal opinion. The conclusions and judgments are based on the experiments.
Translated from Russian by MetaQuotes Ltd.
Original article: https://www.mql5.com/ru/articles/12268
Warning: All rights to these materials are reserved by MetaQuotes Ltd. Copying or reprinting of these materials in whole or in part is prohibited.
This article was written by a user of the site and reflects their personal views. MetaQuotes Ltd is not responsible for the accuracy of the information presented, nor for any consequences resulting from the use of the solutions, strategies or recommendations described.
- Free trading apps
- Over 8,000 signals for copying
- Economic news for exploring financial markets
You agree to website policy and terms of use
The calculation for multivariate variants is rather pessimistic.
In general, yes. That is why usually in multidimensional cases the task of a complete study of the surface device of the loss function is not set. Neither is the task of searching for the global extremum. In fact, one is limited to just finding good enough points. Well, maybe except for cases where it is possible to construct a loss function with good properties, as in MO, for example.
I have a problem with MT5, which does not allow me to access the folder: open Data Folder, in addition to that, I do not have the full elements on the platform MT 5 customizable, I mean by that I do not have the full tools such as rectangle, square or and triangle, etc.. So please, if anyone knows anything about solving these problems, please give me a shout.
you wont get any help by posting on an thread that is unrelated to your post. Create your own thread. And as this is a technical forum, you need to provide more information than you have given including the first 3 lines of the journal when you open mt5 that has your system details.
However, without any more information, it sounds to me like a corrupted installation. try uninstalling mt5, and delete any folders that remain including folders such as "config" that is often not uninstalled correctly. Then try installing it again.