You are missing trading opportunities:
- Free trading apps
- Over 8,000 signals for copying
- Economic news for exploring financial markets
Registration
Log in
You agree to website policy and terms of use
If you do not have an account, please register
It's not a question for me. Is that all you have to say about the article?
What about the article? It's a typical rewrite. It's the same thing in other sources, just in slightly different words. Even the pictures are the same. I didn't see anything new, i.e. authorial.
I wanted to try out the examples, but it's a bummer. The section is for MQL5, but the examples are for MQL4.
vlad1949
Dear Vlad!
Looked through the archives, you have rather old R documentation. It would be good to change to the attached copies.
vlad1949
Dear Vlad!
Why did you fail to run in the tester?
I have everything works without problems. But the scheme is without an indicator: the Expert Advisor communicates directly with R.
Jeffrey Hinton, inventor of deep networks: "Deep networks are only applicable to data where the signal-to-noise ratio is large. Financial series are so noisy that deep networks are not applicable. We've tried it and no luck."
Listen to his lectures on YouTube.
Jeffrey Hinton, inventor of deep networks: "Deep networks are only applicable to data where the signal-to-noise ratio is large. Financial series are so noisy that deep networks are not applicable. We've tried it and no luck."
Listen to his lectures on youtube.
Considering your post in the parallel thread.
Noise is understood differently in classification tasks than in radio engineering. A predictor is considered noisy if it is weakly related (has weak predictive power) for the target variable. A completely different meaning. One should look for predictors that have predictive power for different classes of the target variable.
I have a similar understanding of noise. Financial series depend on a large number of predictors, most of which are unknown to us and which introduce this "noise" into the series. Using only the publicly available predictors, we are unable to predict the target variable no matter what networks or methods we use.
vlad1949
Dear Vlad!
Why did you fail to run in the tester?
I have everything works without problems. True the scheme without an indicator: the advisor directly communicates with R.
llllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllllll
Good afternoon SanSanych.
So the main idea is to make multicurrency with several indicators.
Otherwise, of course, you can pack everything into an Expert Advisor.
But if training, testing and optimisation will be implemented on the fly, without interrupting trading, then the variant with one Expert Advisor will be a bit more difficult to implement.
Good luck
PS. What is the result of testing?
Greetings SanSanych.
Here are some examples of determining the optimal number of clusters that I found on some English-speaking forum. I was not able to use all of them with my data. Very interesting 11 package "clusterSim".
--------------------------------------------------------------------------------------
In the next post calculations with my data
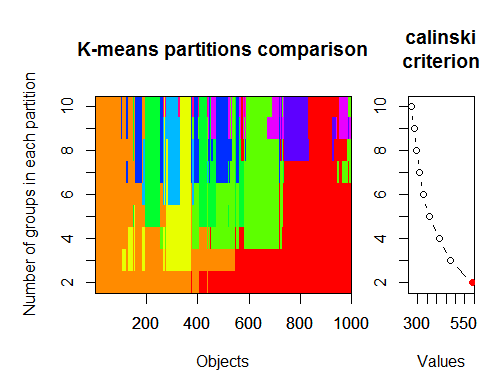
The optimal number of clusters can be determined by several packages and using more than 30 optimality criteria. According to my observations, the most used criterion is the Calinskycriterion .
Let's take the raw data from our set from the indicator dt . It contains 17 predictors, target y and candlestick body z.
In the latest versions of "magrittr" and "dplyr" packages there are many new nice features, one of them is 'pipe' - %>%. It is very convenient when you don't need to save intermediate results. Let's prepare the initial data for clustering. Take the initial matrix dt, select the last 1000 rows from it, and then select 17 columns of our variables from them. We get a clearer notation, nothing more.
1.
2. Calinsky criterion: Another approach to diagnosing how many clusters suit the data. In this case
we try 1 to 10 groups.
3. Determine the optimal model and number of clusters according to the Bayesian InformationCriterion for expectation-maximisation, initialised by hierarchical clustering for parameterised Gaussian mixture models.Gaussian mixture models