Population optimization algorithms: Binary Genetic Algorithm (BGA). Part II
Contents
1. Introduction
2. Algorithm
3. Test results
1. Introduction
In the previous article we got acquainted with all the important basic concepts and methods used not only in genetic algorithms, but, one way or another, in any population optimization algorithms. Now, based on this knowledge, we can consider in detail the main subject of the current "two-volume book" - the binary genetic algorithm (BGA). Starting with general information, let's move on to the code of this remarkable search strategy and finally to the test results.
Genetic algorithm (GA) refers to evolutionary algorithms, which includes various approaches such as genetic algorithms, genetic programming, evolutionary strategies and others. They are based on the ideas of evolution and heredity, where solutions are represented as a population and genetic operators (such as crossover and mutation) are applied to create new generations of solutions.
Genetic algorithms use the principles of natural selection and genetics to solve optimization problems. The binary genetic algorithm (BGA) discussed in the article was the first among all types of genetic algorithms. Thus, BGA belongs to the class of evolutionary algorithms and is a specific variant of a genetic algorithm that uses a binary representation of data.
The work on the creation of genetic algorithms began in the middle of the 20th century. One of the founders is John Holland, who published his book "Adaptation in Natural and Artificial Systems" in 1975, where he introduced the genetic algorithm as a whole direction of approach to solving optimization problems.
The development of the genetic binary algorithm was inspired by several factors and ideas. The main ones:
- Natural selection and principles of evolution: BGA is based on the principles of natural selection and evolution proposed by Charles Darwin. The idea is that there is a diversity of solutions in a population, and those that are better adapted to the environment are more likely to survive and pass on their characteristics to the next generation.
- Genetics and heredity: BGA also uses genetics concepts such as gene, chromosome and heredity. Solutions in BGA are represented as binary strings, where individual groups of bits represent specific genes, and the gene in turn represents the parameter being optimized. Genetic operators, such as crossover and mutation, are applied to binary strings to create new generations of solutions.
Overall, the development of BGA was the result of a combination of ideas from the fields of evolutionary algorithms, genetics and optimization. It was created to solve optimization problems using the principles of natural selection and genetics, and its development continues to this day, a huge number of GA options have been created, as well as the widespread use of ideas and approaches in genetic algorithms as part of hybrid algorithms, including very complex ones.
The Binary Genetic Algorithm (BGA) uses a binary representation of data. This means that each individual (solution) is represented as a string of bits (0 and 1). Genetic operators, such as crossover and mutation, are applied to bit strings to create new generations of solutions.
Interesting fact: Genetic algorithms, including BGA, can be used to create and improve artificial intelligence. For example, they can be used to evolve neural networks, allowing the creation of more efficient machine learning models.
Genetic algorithms in general and BGA in particular are a powerful tool for solving complex optimization problems where traditional methods may not be effective enough due to the lack of analytical solution. MetaTrader 5 uses a binary GA and therefore it is even more exciting to study the operation principles of this remarkable algorithm.
2. Algorithm
The binary genetic algorithm includes the following steps:
- Population initialization - create an initial population consisting of chromosomes with random binary values.
- Fitness assessment - evaluate the fitness of each individual (chromosome) in the daughter population.
- Selection - select parents to create offspring using the roulette wheel method.
- Crossover - break the parent chromosomes into sections and create new daughter chromosomes with sections of both parents.
- Inversion - split the chromosome of a daughter individual at a randomly selected point and swap the resulting sections.
- Mutation - randomly change bits in the genes of the offspring with a given probability of mutation.
- Assessment of offspring fitness - assess the fitness of each new offspring.
- Forming a new population - place the offspring population at the end of the total population and sort by fitness value.
- Stopping criterion - repeat the process from step 3 until the stopping criterion is reached.
We will use the distance between the "max" and "min" of the optimized parameters to ensure that the BGA works only with positive numbers, simplify operations with binary strings and increase the speed of calculations. We will represent the positive distance values obtained in this way in the form of a binary Gray code and place them sequentially in one common array of chromosome symbols as shown in Figure 1. When performing the crossover method, chromosome breakpoints are located in random places on the chromosome and not necessarily at gene joining points. Breakpoints can also be inside the gene space.
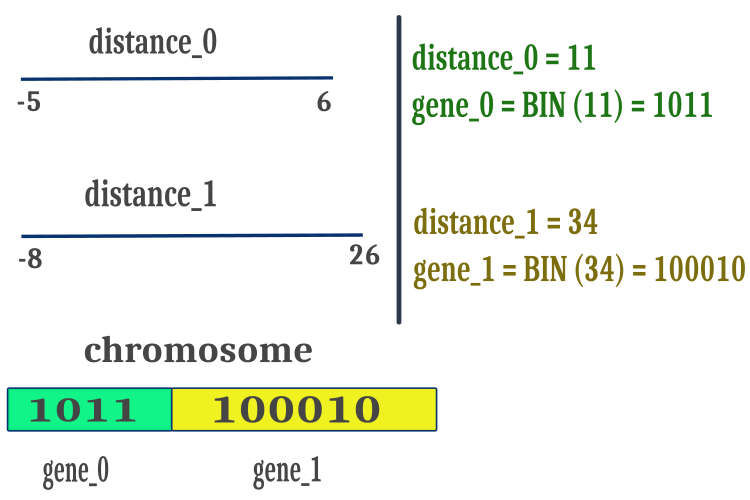
Figure 1. Placing the characteristics of an individual (optimized parameters) in the chromosome
There are a significant number of external parameters for a genetic algorithm, so it is reasonable to consider them in more detail. The default settings are almost entirely consistent with the recommendations of many authors of scientific publications. I have tested and selected them to provide maximum efficiency in most types of tasks. However, deviation from these parameters in any direction can lead to achieving 100% convergence on tests with 10 or even 50 optimized parameters on individual functions, but at the same time significantly reduce efficiency on other tasks. Therefore, the default settings are optimal for use in most practical cases.
- Population_P (population size = 50) - the number of daughter individuals in each generation of the population. This population size is used in most of the algorithms covered in the tests and provides a balance between solution diversity and convergence speed.
- ParentPopulation_P (parent population size = 50) - the number of parents selected for breeding and creating the next generation. Decreasing the value of this parameter improves convergence on smooth functions (increases accuracy), increasing the value improves convergence on discrete functions (increases the diversity of genetic material).
- CrossoverProbab_P (crossover probability = 1.0) - the probability of performing a crossover operation between two selected parents. A high probability of crossover increases the combinatorial capabilities of the algorithm. Decreasing the value increases the speed of convergence, but increases the probability of getting stuck.
- CrossoverPoints_P (number of crossover points = 3) - the number of points, at which a crossover between the two parent chromosomes occurs. Increasing the points leads to more intense mixing of parameters among themselves and, in the limit, reduces to the algorithm random behavior.
- MutationProbab_P (mutation probability = 0.001) - the probability of mutation of each bit in the chromosome genotype. Mutation allows random changes to be made to genetic material and adds new solutions to a population. A low probability increases the convergence rate of the algorithm but reduces diversity, while a too high probability results in the loss of useful information.
- InversionProbab_P (inversion probability = 0.7) - the probability of performing an inversion operation on a chromosome. A high probability of inversion increases the diversity of the genetic material, but too high a probability leads to the algorithm random behavior.
- DoubleDigitsInChromo_P (number of decimal places in the gene) - the number of decimal places of the real number (optimized parameter) represented in binary form in the chromosome. Increasing the value leads to an increase in the complexity of calculations and an increase in optimization time (without using the binary form directly in solving the problem, it makes no sense to set more than 16 - the output bits will be lost when converting to double).
Let's move on to reviewing the code.
When initializing agents, it is necessary to determine the number of bits in the binary representation of the parameter being optimized. Let's say we need to consider five decimal places, which corresponds to a certain Gray code length. However, more than the required number may be encoded in this code. Therefore, we need to determine the maximum real number that can be encoded in the binary format. In the future, we can scale the encoded number to the required range of the optimized output parameter. For the fractional part of a real number, we use a given number of digits (specified in the parameters), and for the integer part - as many as required in binary form.
For example, if the input parameter of the digitsInGrayCode function is 3, then the function will return the maximum value of ulong type using the Gray code for 3 bits, that is, 7 (111 in binary).
In order to determin the maximum possible number that can be encoded by a given number of bits for the fractional and integer part of a real number, we will use the GetMaxDecimalFromGray function.
//—————————————————————————————————————————————————————————————————————————————— //Calculation of the maximum possible ulong number using the Gray code for a given number of bits ulong GetMaxDecimalFromGray (int digitsInGrayCode) { ulong maxValue = 1; for (int i = 1; i < digitsInGrayCode; i++) { maxValue <<= 1; maxValue |= 1; } return maxValue; } //——————————————————————————————————————————————————————————————————————————————
To represent each gene in the chromosome (the position of the gene on the chromosome), we will use the S_BinaryGene structure, which contains fields and methods for working with genes in binary format:
- "gene" - character array representing a gene.
- "integPart" - character array of the integer gene part.
- "fractPart" - character array of the fractional gene part.
- "integGrayDigits" - number of Gray digits for the integer part of the gene.
- "fractGrayDigits" - number of Gray digits for the fractional part of the gene.
- "length" - total gene length.
- "minDoubleRange" - minimum value of the real number range.
- "maxDoubleRange" - maximum value of the real number range.
- "maxDoubleNumber" - maximum real number that can be represented using the gene.
- "doubleFract" - value to convert the fractional part of the gene to a real number.
Structure methods:
- "Init" - initializes the structure fields. Accepts the minimum and maximum values of a range of real numbers, as well as the number of digits in the fractional part of the gene. Within the method, values for the maximum real number of coding parts of the gene are calculated using Gray code.
- "ToDouble" - converts the gene to a real number. Accepts the "chromo" (chromosome) array of Gray code characters and the "indChr" gene start index. The method reads a region of a chromosome, converts the read gene into a decimal value and then scales it to a specified range of real numbers.
- "Scale" - scales the "In" input value from the "InMIN" and "InMAX" range to the "OutMIN" and "OutMAX" output range. This is an auxiliary method used in "ToDouble".
//—————————————————————————————————————————————————————————————————————————————— struct S_BinaryGene { char gene []; char integPart []; char fractPart []; uint integGrayDigits; uint fractGrayDigits; uint length; double minDoubleRange; double maxDoubleRange; double maxDoubleNumber; double doubleFract; //---------------------------------------------------------------------------- void Init (double min, double max, int doubleDigitsInChromo) { doubleFract = pow (0.1, doubleDigitsInChromo); minDoubleRange = min; maxDoubleRange = max - min; ulong decInfr = 0; for (int i = 0; i < doubleDigitsInChromo; i++) { decInfr += 9 * (ulong)pow (10, i); } //---------------------------------------- DecimalToGray (decInfr, fractPart); ulong maxDecInFr = GetMaxDecimalFromGray (ArraySize (fractPart)); double maxDoubFr = maxDecInFr * doubleFract; //---------------------------------------- DecimalToGray ((ulong)maxDoubleRange, integPart); ulong maxDecInInteg = GetMaxDecimalFromGray (ArraySize (integPart)); double maxDoubInteg = (double)maxDecInInteg + maxDoubFr; maxDoubleNumber = maxDoubInteg; ArrayResize (gene, 0, 1000); integGrayDigits = ArraySize (integPart); fractGrayDigits = ArraySize (fractPart); length = integGrayDigits + fractGrayDigits; ArrayCopy (gene, integPart, 0, 0, WHOLE_ARRAY); ArrayCopy (gene, fractPart, ArraySize (gene), 0, WHOLE_ARRAY); } //---------------------------------------------------------------------------- double ToDouble (const char &chromo [], const int indChr) { double d; if(integGrayDigits > 0)d = (double)GrayToDecimal(chromo, indChr, indChr + integGrayDigits - 1); else d = 0.0; d +=(double)GrayToDecimal(chromo, indChr + integGrayDigits, indChr + integGrayDigits + fractGrayDigits - 1) * doubleFract; return minDoubleRange + Scale(d, 0.0, maxDoubleNumber, 0.0, maxDoubleRange); } //---------------------------------------------------------------------------- double Scale (double In, double InMIN, double InMAX, double OutMIN, double OutMAX) { if (OutMIN == OutMAX) return (OutMIN); if (InMIN == InMAX) return (double((OutMIN + OutMAX) / 2.0)); else { if (In < InMIN) return OutMIN; if (In > InMAX) return OutMAX; return (((In - InMIN) * (OutMAX - OutMIN) / (InMAX - InMIN)) + OutMIN); } } }; //——————————————————————————————————————————————————————————————————————————————
To describe the agent, we need the S_Agent structure, which represents the agent and contains the following data in addition to the chromosome:
- "c" - array of agent coordinate values as a real number.
- "f" - agent fitness value.
- "genes" - array of "S_BinaryGene" structures that describe the position of each gene in the chromosome and the rules for converting the binary code into a real number.
- "chromosome" - array of agent chromosome characters.
- "calculated" - a boolean value indicating whether the agent's values have been calculated (the field is present, but not used in the code).
Structure methods:
- "Init" - initializes the structure fields. Accepts the number of "coords" coordinates, "min" and "max" arrays with minimum and maximum values for each optimized parameter, as well as doubleDigitsInChromo - the number of digits of a real number in the fractional part of the gene. The method creates and initializes genes for each coordinate, and also sets the initial fitness value and the "calculated" flag.
- "ExtractGenes" - extracts gene values from a chromosome and stores them in "c" array, uses the "ToDouble" method from the "S_BinaryGene" structure to convert genes from a chromosome to real numbers.
//—————————————————————————————————————————————————————————————————————————————— struct S_Agent { void Init (const int coords, const double &min [], const double &max [], int doubleDigitsInChromo) { ArrayResize(c, coords); f = -DBL_MAX; ArrayResize(genes, coords); ArrayResize(chromosome, 0, 1000); for(int i = 0; i < coords; i++) { genes [i].Init(min [i], max [i], doubleDigitsInChromo); ArrayCopy(chromosome, genes [i].gene, ArraySize(chromosome), 0, WHOLE_ARRAY); } calculated = false; } void ExtractGenes () { uint pos = 0; for (int i = 0; i < ArraySize (genes); i++) { c [i] = genes [i].ToDouble (chromosome, pos); pos += genes [i].length; } } double c []; //coordinates double f; //fitness S_BinaryGene genes []; char chromosome []; bool calculated; }; //——————————————————————————————————————————————————————————————————————————————
The following code presents the definition of the S_Roulette structure, which represents the roulette segment.
Structure fields:
- "start" - roulette segment starting point value.
- "end" - roulette segment end point.
//—————————————————————————————————————————————————————————————————————————————— struct S_Roulette { double start; double end; }; //——————————————————————————————————————————————————————————————————————————————
Declare the C_AO_BGA class that implements the genetic algorithm:
- "cB" - array of best coordinate values.
- "fB" - fitness value of the best coordinates.
- "a" - array of S_Agent structures representing agents.
- "rangeMax" - array of maximum search range values.
- "rangeMin" - array of minimum search range values.
- "rangeStep" - array of values representing the search step.
Class methods:
- "Init" - initializes the class field. It accepts the following parameters: "coordsP" number of coordinates, "popSizeP" population size, "parentPopSizeP" parent population size, "crossoverProbabP" crossing probability, "crossoverPointsP" number of crossing points, "mutationProbabP" mutation probability, "inversionProbabP" inversion probability, "doubleDigitsInChromoP" number of decimal places in the gene. The method initializes internal variables and arrays necessary for the operation of the genetic algorithm.
- "Moving" - main method of the genetic algorithm, performs operations on the population, such as crossover, mutation, inversion and fitness assessment.
- "Revision" - the method performs a population revision, sorting agents and selecting the best.
The class's private fields and methods are used to implement the genetic algorithm internally, including roulette operations, value scaling, sorting and other operations.
//—————————————————————————————————————————————————————————————————————————————— class C_AO_BGA { //---------------------------------------------------------------------------- public: double cB []; //best coordinates public: double fB; //FF of the best coordinates public: S_Agent a []; //agent public: double rangeMax []; //maximum search range public: double rangeMin []; //manimum search range public: double rangeStep []; //step search public: void Init (const int coordsP, //coordinates number const int popSizeP, //population size const int parentPopSizeP, //parent population size const double crossoverProbabP, //crossover probability const int crossoverPointsP, //crossover points const double mutationProbabP, //mutation probability const double inversionProbabP, //inversion probability const int doubleDigitsInChromoP);//number of decimal places in the gene public: void Moving (); public: void Revision (); //---------------------------------------------------------------------------- private: int coords; //coordinates number private: int popSize; //population size private: int parentPopSize; //parent population size private: double crossoverProbab; //crossover probability private: int crossoverPoints; //crossover points private: double mutationProbab; //mutation probability private: double inversionProbab; //inversion probability private: int doubleDigitsInChromo; //number of decimal places in the gene private: bool revision; private: S_Agent parents []; //parents private: int ind []; //temporary array for sorting the population private: double val []; //temporary array for sorting the population private: S_Agent pTemp []; //temporary array for sorting the population private: char tempChrome []; //temporary chromosome for inversion surgery private: uint lengthChrome; //length of the chromosome (the length of the string of characters according to the Gray code) private: int pCount; //indices of chromosome break points private: uint poRND []; //temporal indices of chromosome break points private: uint points []; //final indices of chromosome break points private: S_Roulette roulette []; //roulette private: void PreCalcRoulette (); private: int SpinRoulette (); private: double SeInDiSp (double In, double InMin, double InMax, double Step); private: double RNDfromCI (double min, double max); private: void Sorting (S_Agent &p [], int size); private: double Scale (double In, double InMIN, double InMAX, double OutMIN, double OutMAX); }; //——————————————————————————————————————————————————————————————————————————————
The following code represents the implementation of the Init method of the C_AO_BGA class. This method initializes the class fields and arrays necessary for the genetic algorithm to work.
Method inputs:
- "coordsP" - number of coordinates.
- "popSizeP" - population size.
- "parentPopSizeP" - parent population size.
- "crossoverProbabP" - crossover probability.
- "crossoverPointsP" - number of crossover points.
- "mutationProbabP" - mutation probability.
- "inversionProbabP" - inversion probability.
- "doubleDigitsInChromoP" - number of decimal places in the gene.
The Init method is used to initialize class fields and arrays before executing the genetic algorithm. It sets the values of class fields, checks and adjusts the values of some parameters and also resizes arrays by allocating memory for them.
//—————————————————————————————————————————————————————————————————————————————— void C_AO_BGA::Init (const int coordsP, //coordinates number const int popSizeP, //population size const int parentPopSizeP, //parent population size const double crossoverProbabP, //crossover probability const int crossoverPointsP, //crossover points const double mutationProbabP, //mutation probability const double inversionProbabP, //inversion probability const int doubleDigitsInChromoP) //number of decimal places in the gene { MathSrand ((int)GetMicrosecondCount ()); // reset of the generator fB = -DBL_MAX; revision = false; coords = coordsP; popSize = popSizeP; parentPopSize = parentPopSizeP; crossoverProbab = crossoverProbabP; crossoverPoints = crossoverPointsP; pCount = crossoverPointsP; mutationProbab = mutationProbabP; inversionProbab = inversionProbabP; doubleDigitsInChromo = doubleDigitsInChromoP; if (crossoverPoints < 1) crossoverPoints = 1; if (pCount < 1) pCount = 1; ArrayResize (poRND, pCount); ArrayResize (points, pCount + 2); ArrayResize (ind, parentPopSize + popSize); ArrayResize (val, parentPopSize + popSize); ArrayResize (pTemp, parentPopSize + popSize); ArrayResize (a, popSize); ArrayResize (parents, parentPopSize + popSize); ArrayResize (roulette, parentPopSize); ArrayResize (rangeMax, coords); ArrayResize (rangeMin, coords); ArrayResize (rangeStep, coords); ArrayResize (cB, coords); } //——————————————————————————————————————————————————————————————————————————————
All basic operations for working with the genetic material of agents are performed using the Moving method of the C_AO_BGA class. The Moving method performs a genetic algorithm step, including selecting parents, crossover, inverting and mutating chromosomes and applying operations to genes and coordinates of individuals.
The method is logically divided into several parts:
1. "if (!revision)" - if "revision" is equal to "false", the population individuals are initialized:
- The Init method is called to initialize the individual with the given parameters.
- A random chromosome is generated by filling each gene with a random value of 0 or 1.
- The ExtractGenes method is called to extract genes from the chromosome.
- The coordinates of "c" individual are reduced to a range using the SeInDiSp function.
- The fitness value of each individual "f" is set to "-DBL_MAX".
- "lengthChrome = ArraySize (a [0].chromosome)" - length of an individual's chromosome (all individuals have the same length).
- "ArrayResize (tempChrome, lengthChrome)" - replaces the size of the "tempChrome" temporary array with "lengthChrome".
2. For each individual in the population:
- A preliminary calculation of the parent selection roulette is performed using the PreCalcRoulette method.
- The parent is selected using the SpinRoulette method.
- The chromosome of the selected parent individual is copied into the chromosome of the current individual.
- The crossing operation is performed with the crossoverProbab probability.
- The second parent is selected.
- Chromosome breakpoints are determined.
- The chromosomes of the parent individuals are crossed.
- The inversion operation is performed with the inversionProbab probability.
- A random chromosome breakpoint is determined.
- Parts of the chromosome change places.
- A mutation operation is performed for each chromosome gene with the mutationProbab probability.
- The ExtractGenes method is called to extract genes from the chromosome.
- The coordinates of "c" individual are reduced to a range using the SeInDiSp function.
//—————————————————————————————————————————————————————————————————————————————— void C_AO_BGA::Moving () { //---------------------------------------------------------------------------- if (!revision) { for (int i = 0; i < popSize; i++) { a [i].Init (coords, rangeMin, rangeMax, doubleDigitsInChromo); int r = 0; for (int len = 0; len < ArraySize (a [i].chromosome); len++) { r = MathRand (); //[0,32767] if (r > 16384) a [i].chromosome [len] = 1; else a [i].chromosome [len] = 0; } a [i].ExtractGenes (); for (int c = 0; c < coords; c++) a [i].c [c] = SeInDiSp (a [i].c [c], rangeMin [c], rangeMax [c], rangeStep [c]); a [i].f = -DBL_MAX; a [i].calculated = true; } lengthChrome = ArraySize (a [0].chromosome); ArrayResize (tempChrome, lengthChrome); for (int i = 0; i < parentPopSize + popSize; i++) { parents [i].Init (coords, rangeMin, rangeMax, doubleDigitsInChromo); parents [i].f = -DBL_MAX; } revision = true; return; } //---------------------------------------------------------------------------- int pos = 0; double r = 0; uint p1 = 0; uint p2 = 0; uint p3 = 0; uint temp = 0; for (int i = 0; i < popSize; i++) { PreCalcRoulette (); //selection, select and copy the parent to the child------------------------ pos = SpinRoulette (); ArrayCopy (a [i].chromosome, parents [pos].chromosome, 0, 0, WHOLE_ARRAY); //crossover----------------------------------------------------------------- r = RNDfromCI (0.0, 1.0); if (r < crossoverProbab) { //choose a second parent to breed with------------------------------------ pos = SpinRoulette (); //determination of chromosome break points-------------------------------- for (int p = 0; p < pCount; p++) { poRND [p] = (int)RNDfromCI (0.0, lengthChrome); if (poRND [p] >= lengthChrome) poRND [p] = lengthChrome - 1; } ArraySort (poRND); ArrayCopy (points, poRND, 1, 0, WHOLE_ARRAY); points [0] = 0; points [pCount + 1] = lengthChrome - 1; r = RNDfromCI (0.0, 1.0); int startPoint = r > 0.5 ? 0 : 1; for (int p = startPoint; p < pCount + 2; p += 2) { if (p < pCount + 1) { for (uint len = points [p]; len < points [p + 1]; len++) a [i].chromosome [len] = parents [pos].chromosome [len]; } } } //perform an inversion------------------------------------------------------ //(break the chromosome, swap the received parts, connect them together) r = RNDfromCI (0.0, 1.0); if (r < inversionProbab) { p1 = (int)RNDfromCI (0.0, lengthChrome); if (p1 >= lengthChrome) p1 = lengthChrome - 1; //copying the second part to the beginning of the temporary array for (uint len = p1; len < lengthChrome; len++) tempChrome [len - p1] = a [i].chromosome [len]; //copying the first part to the end of the temporary array for (uint len = 0; len < p1; len++) tempChrome [lengthChrome - p1 + len] = a [i].chromosome [len]; //copying a temporary array back for (uint len = 0; len < lengthChrome; len++) a [i].chromosome [len] = tempChrome [len]; } //perform mutation--------------------------------------------------------- for (uint len = 0; len < lengthChrome; len++) { r = RNDfromCI (0.0, 1.0); if (r < mutationProbab) a [i].chromosome [len] = a [i].chromosome [len] == 1 ? 0 : 1; } a [i].ExtractGenes (); for (int c = 0; c < coords; c++) a [i].c [c] = SeInDiSp (a [i].c [c], rangeMin [c], rangeMax [c], rangeStep [c]); a [i].calculated = true; } } //——————————————————————————————————————————————————————————————————————————————
The Revision method performs a population revision, sorts individuals by the value of their fitness function and updates the "fB" fitness of the best solution and the "cB" coordinates of the best solution. Before sorting the population, the child population is copied to the end of the total population.
//—————————————————————————————————————————————————————————————————————————————— void C_AO_BGA::Revision () { //---------------------------------------------------------------------------- for (int i = parentPopSize; i < parentPopSize + popSize; i++) { parents [i] = a [i - parentPopSize]; } Sorting (parents, parentPopSize + popSize); if (parents [0].f > fB) { fB = parents [0].f; ArrayCopy (cB, parents [0].c, 0, 0, WHOLE_ARRAY); } } //——————————————————————————————————————————————————————————————————————————————
The preliminary roulette layout code represents the PreCalcRoulette method. This method preliminarily computes range values for a roulette selection of individuals based on their fitness function.
//—————————————————————————————————————————————————————————————————————————————— void C_AO_BGA::PreCalcRoulette () { roulette [0].start = parents [0].f; roulette [0].end = roulette [0].start + (parents [0].f - parents [parentPopSize - 1].f); for (int s = 1; s < parentPopSize; s++) { if (s != parentPopSize - 1) { roulette [s].start = roulette [s - 1].end; roulette [s].end = roulette [s].start + (parents [s].f - parents [parentPopSize - 1].f); } else { roulette [s].start = roulette [s - 1].end; roulette [s].end = roulette [s].start + (parents [s - 1].f - parents [s].f) * 0.1; } } } //——————————————————————————————————————————————————————————————————————————————
The SpinRoulette method ensures the position of the parent individual is selected.
//—————————————————————————————————————————————————————————————————————————————— int C_AO_BGA::SpinRoulette () { double r = RNDfromCI (roulette [0].start, roulette [parentPopSize - 1].end); for (int s = 0; s < parentPopSize; s++) { if (roulette [s].start <= r && r < roulette [s].end) return s; } return 0; } //——————————————————————————————————————————————————————————————————————————————
3. Test results
BGA test stand results:
C_AO_BGA:50:50:1.0:3:0.001:0.7:3
=============================
5 Hilly's; Func runs: 10000; result: 0.9999191151339382
25 Hilly's; Func runs: 10000; result: 0.994841435673127
500 Hilly's; Func runs: 10000; result: 0.5048331764136147
=============================
5 Forest's; Func runs: 10000; result: 1.0
25 Forest's; Func runs: 10000; result: 0.9997457419655973
500 Forest's; Func runs: 10000; result: 0.32054251149158375
=============================
5 Megacity's; Func runs: 10000; result: 0.9066666666666668
25 Megacity's; Func runs: 10000; result: 0.9640000000000001
500 Megacity's; Func runs: 10000; result: 0.23034999999999997
=============================
All score: 6.92090 (76.90%)
Visualization of the BGA optimization clearly demonstrates the high convergence of the algorithm. Interestingly, during the optimization process, some part of the population remains away from the global extremum, which indicates the exploration of new unknown regions of the search space, maintaining the diversity of solutions in the population. However, the algorithm faces certain difficulties when working with the Megacity discrete function, which is problematic for most algorithms. Despite some variation in results when working with this complex function, BGA performs significantly better than other algorithms.
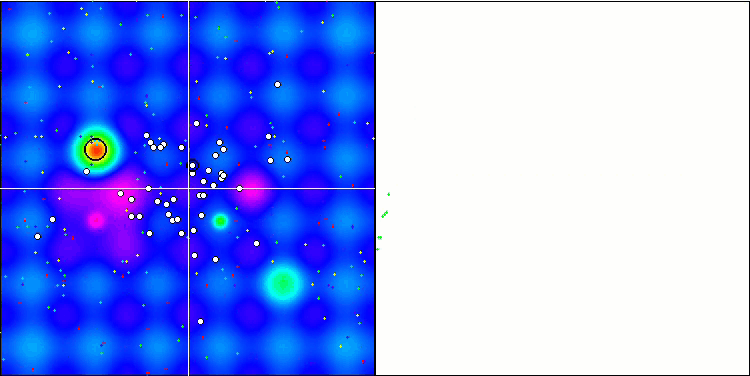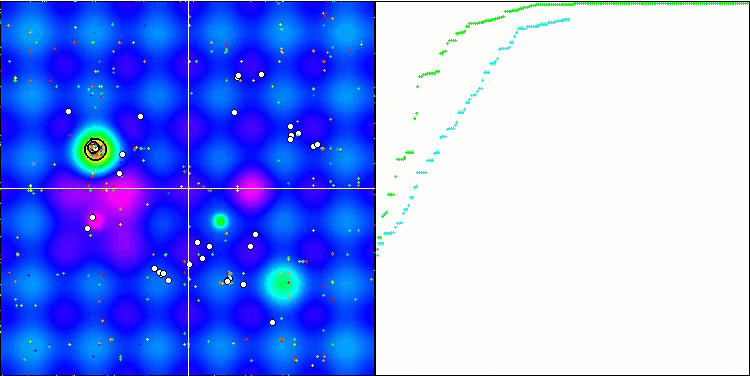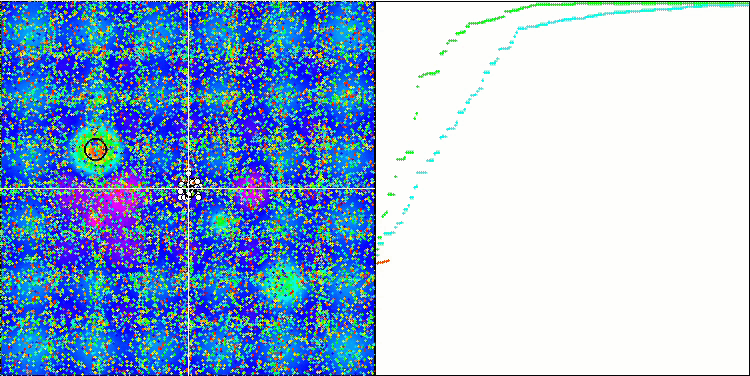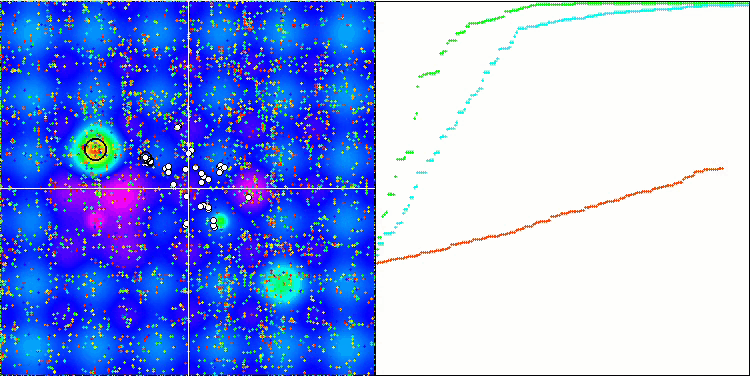
BGA on the Hilly test function
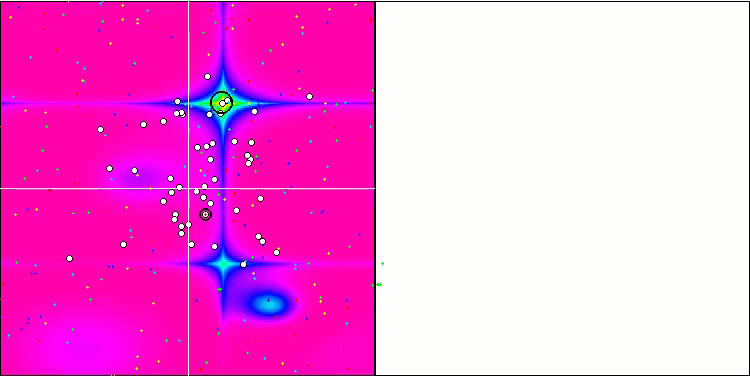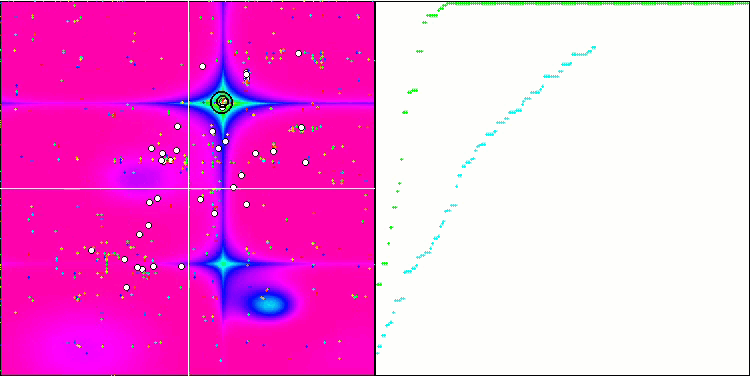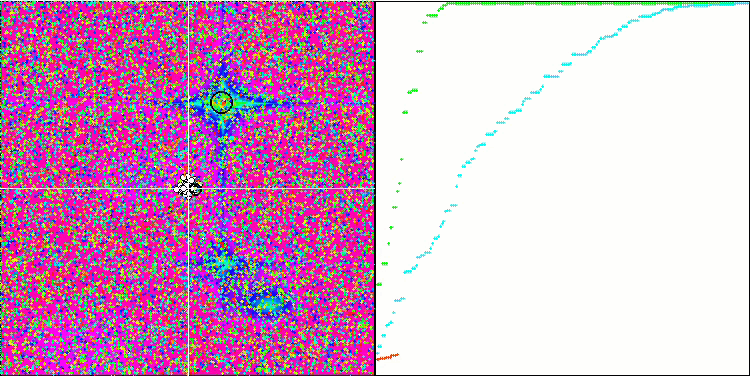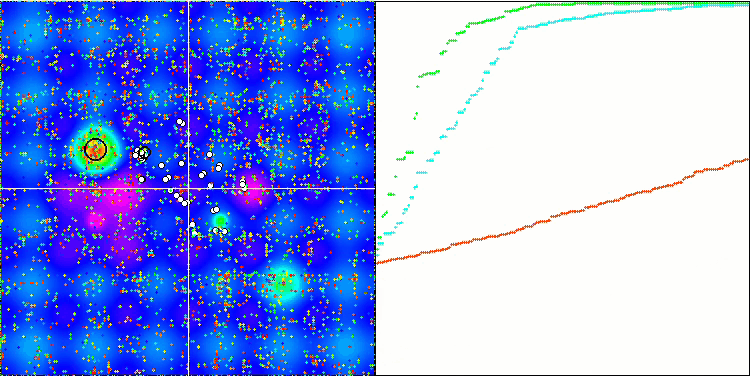
BGA on the Forest test function
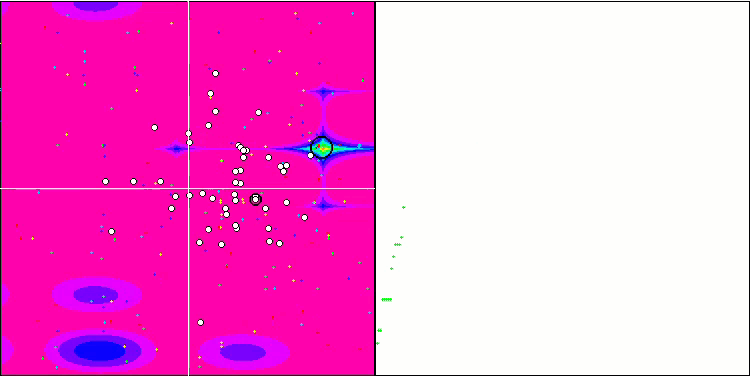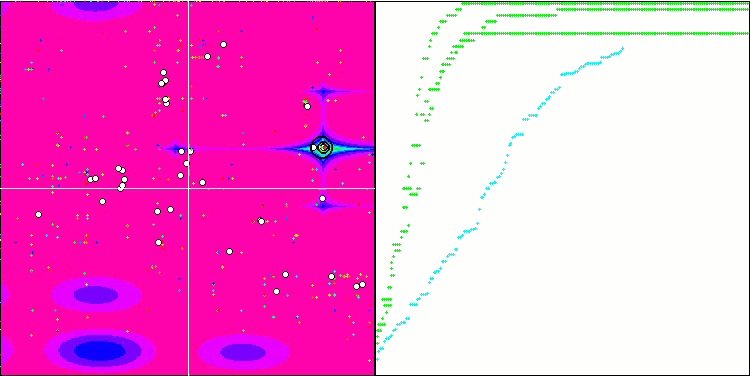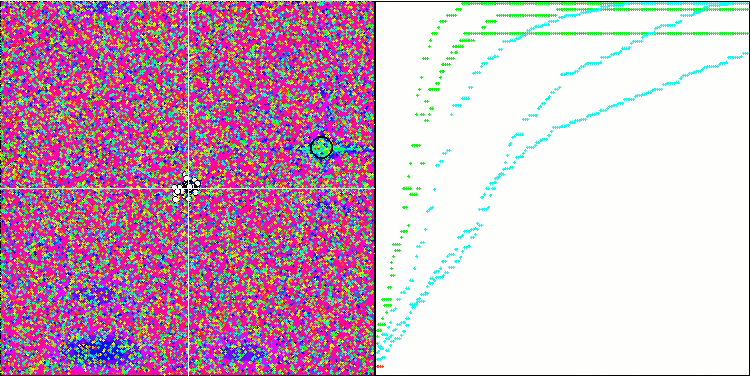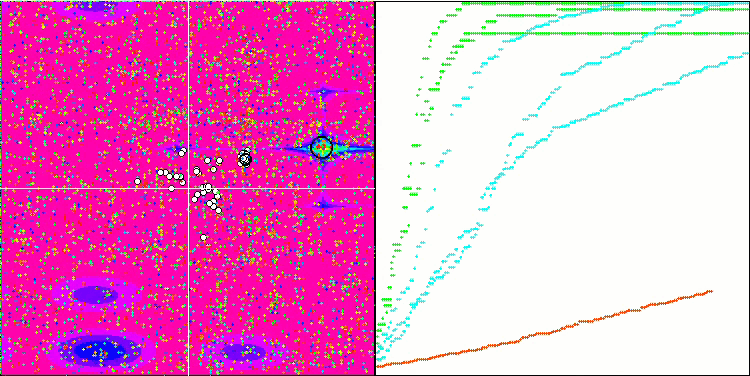
BGA on the Megacity test function
BGA took the top spot in the table, performing best in most tests for all three test functions. BGA demonstrated especially impressive results on the Megacity discrete function, outperforming other algorithms.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Summary
In this article, we examined the classic version of BGA, a special case of the general class of GA genetic algorithms, and all conclusions relate specifically to it. Despite the long-standing idea of representing solutions in binary, the approach using binary code remains relevant to this day. It combines the independent spatial dimensions of an optimization problem into a single whole by encoding information in a single chromosome, which is difficult to implement using conventional real-valued feature encoding, making this algorithm stand out among other optimization algorithms.
Although the mathematics and logic of the BGA strategy are completely clear to me, I am still fascinated by what is happening with the chromosome. It can be compared to a magical kaleidoscope. As we spin the kaleidoscope, the varied shapes and colors combine into unique patterns to create a spectacular picture. Similarly, the crossover operator in BGA randomly cuts the chromosome into several parts, including internal parameter areas. These pieces are then put together, like shuffling the pieces of a kaleidoscope. This allows us to combine the best features of different solutions and create a new, more optimal combination. Just like a kaleidoscope, the results of BGA crossover can be surprising and unexpected, turning simple chromosomes into true "diamonds" of optimal solutions.
I am confident that the information in this two-part article on the methods and tools used in genetic algorithms will help you expand your knowledge and reach new heights in your work and research. The power of evolution is constantly manifesting itself in nature, technology and the human mind, and BGA is one of many amazing algorithms that will help us reach new heights and achievements.
BGA effectively solves a variety of problems, be it smooth surfaces, discrete problems or even large-scale problems, including a stochastic approach, opening up new possibilities where analytical solutions are limited.
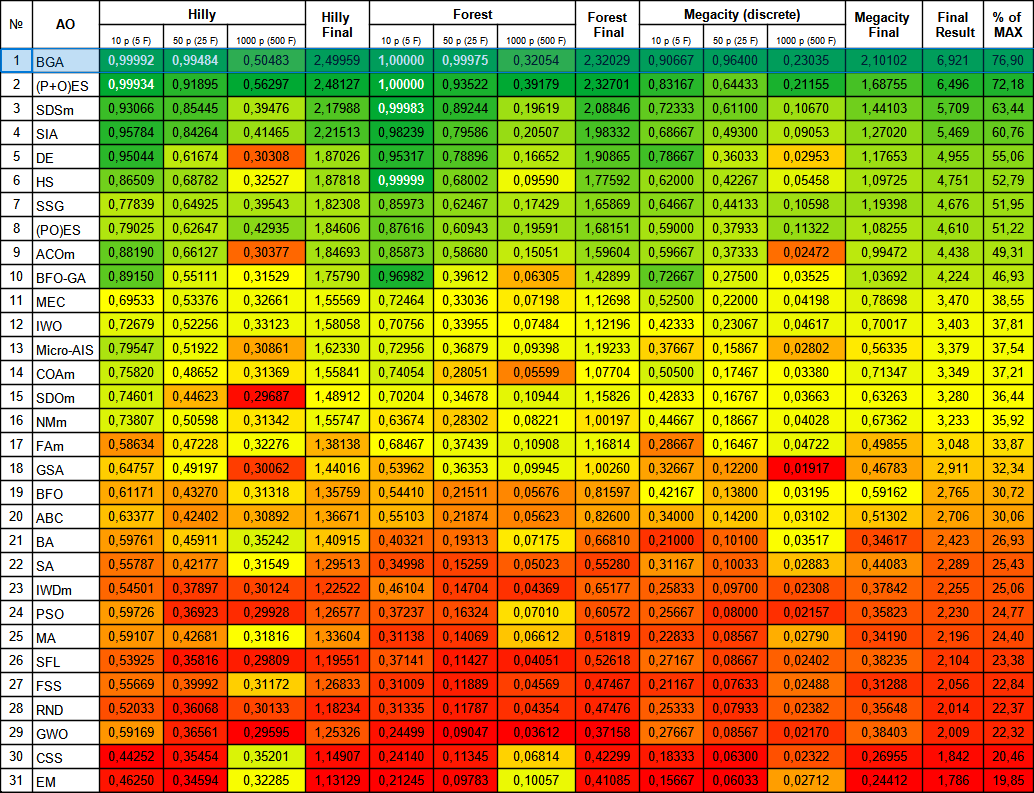
Figure 2. Color gradation of algorithms according to relevant tests Results greater than or equal to 0.99 are highlighted in white
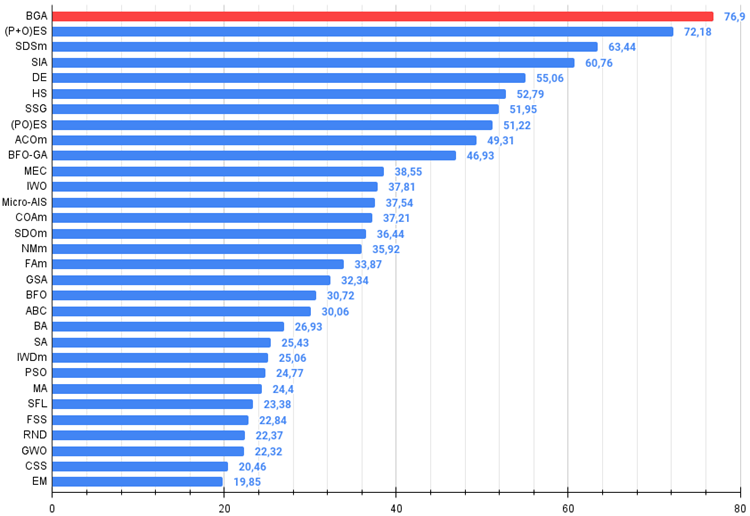
Figure 3. The histogram of algorithm test results (on a scale from 0 to 100, the more the better,
where 100 is the maximum possible theoretical result, the archive features a script for calculating the rating table)
The pros and cons of the binary genetic algorithm (BGA) apply exclusively to the presented implementation:
Advantages:
- High efficiency in solving a variety of problems.
- Resistance to sticking.
- Promising results on both smooth and complex discrete functions.
- High convergence.
Disadvantages:
- A large number of external parameters.
- Quite a complex implementation.
- High computational complexity.
The article is accompanied by an archive with updated current versions of the algorithm codes described in previous articles. The author of the article is not responsible for the absolute accuracy in the description of canonical algorithms. Changes have been made to many of them to improve search capabilities. The conclusions and judgments presented in the articles are based on the results of the experiments.
Translated from Russian by MetaQuotes Ltd.
Original article: https://www.mql5.com/ru/articles/14040
 MQL5 Wizard Techniques you should know (Part 20): Symbolic Regression
MQL5 Wizard Techniques you should know (Part 20): Symbolic Regression
 Learn how to trade the Fair Value Gap (FVG)/Imbalances step-by-step: A Smart Money concept approach
Learn how to trade the Fair Value Gap (FVG)/Imbalances step-by-step: A Smart Money concept approach
 Developing a multi-currency Expert Advisor (Part 1): Collaboration of several trading strategies
Developing a multi-currency Expert Advisor (Part 1): Collaboration of several trading strategies
 Triangular arbitrage with predictions
Triangular arbitrage with predictions
- Free trading apps
- Over 8,000 signals for copying
- Economic news for exploring financial markets
You agree to website policy and terms of use